Eukaryotic chromatin is composed of nucleosomes that form a bead-like structure and are further folded and packed. Transcription of genes requires that this higher structure be unpacked into a more loosely structured chromatin. This loose structure makes the regulatory elements (promoters, enhancers, etc.) on the DNA physically accessible. It thus can be bound by proteins such as transcription factors, activating the transcription process. ATAC-seq (assay for transposase-accessible chromatin with high throughput sequencing) is an epigenetic research technique for sequencing and analyzing the accessible nuclear chromatin region of Tn5 transposase.
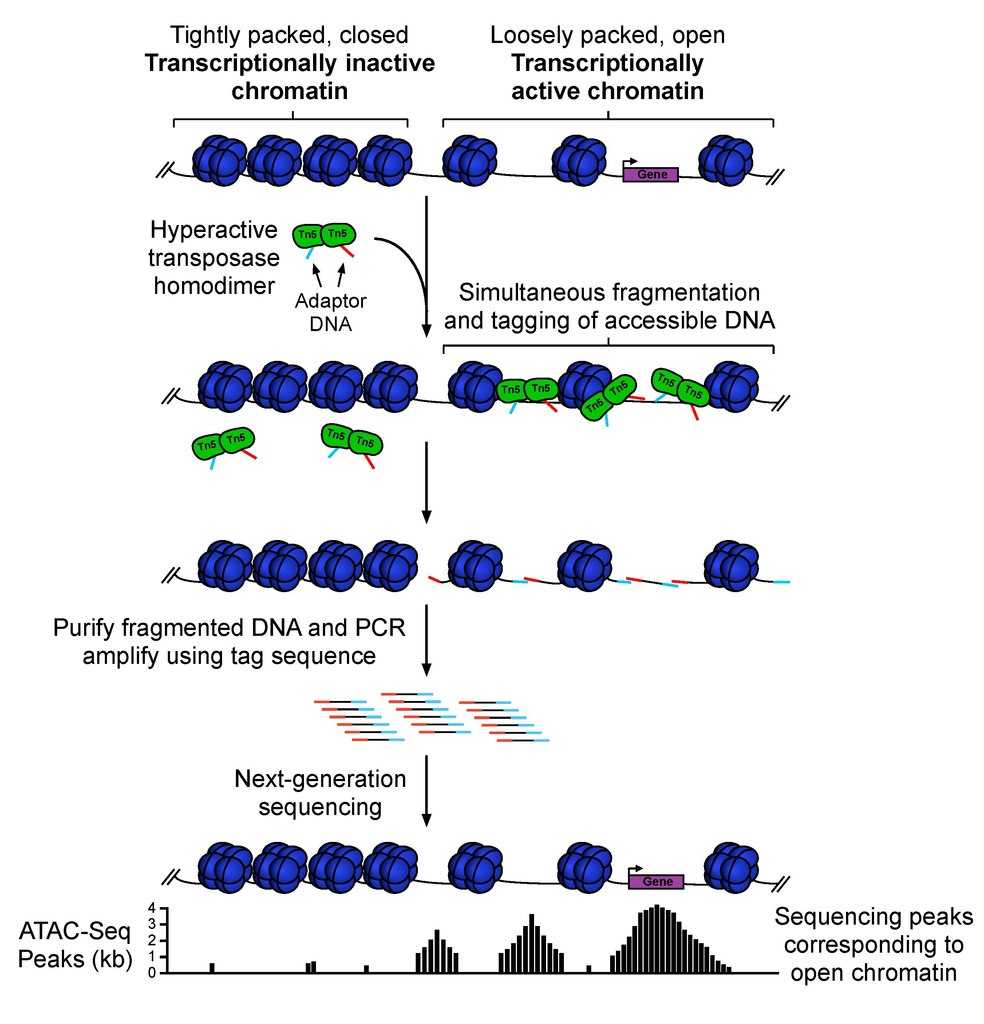 Figure 1. Schematic diagram of the main steps of ATAC-Seq. (Grandi, F. C, et al. 2022)
Figure 1. Schematic diagram of the main steps of ATAC-Seq. (Grandi, F. C, et al. 2022)
Lifeasible uses Tn5 transposase to cleave the nuclear chromatin region developed at a specific time and space to obtain all actively transcribed regulatory sequences in the genome at that time and space and then clusters these sequences to mine potential binding transcription factors at these open loci by motif analysis, which is combined with gene expression level data to discover critical regulatory transcription factors.
Lifeasible's ATAC-seq program has the advantage of "short cycle time, wide range, and high specificity" and has been widely used in the study of mice, rats, pigs, bovine, equines, fish, marine invertebrates, non-human primates, insects, and other species. It is our ideal to meet all the needs of our customers and to provide accurate and true data. We look forward to working with you; please feel free to contact us.
Reference