The Pleiotropic drug resistance (PDR) transporter protein in plants is widely involved in nutrient uptake, developmental regulation, detoxification processes, and plant defense responses to biotic and abiotic stresses. Recent studies have also shown that this protein is associated with resistance to various fungal pathogens in plants. With the rapid development of plant genome sequencing technology, more and more scientists are mining, comparative analysis, and functional prediction of plant PDR at the genome level.
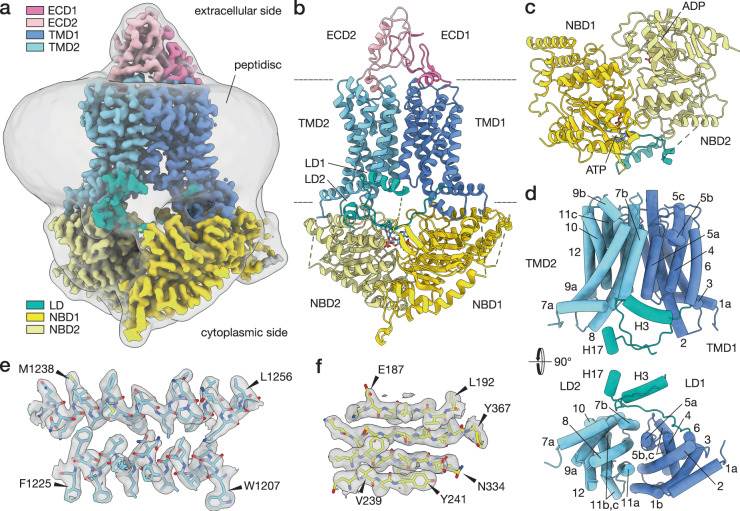 Figure 1. Cryo-electron micrograph of the inwardly open conformation of PDR5 (ADP-PDR5). (Harris, A et al., 2021)
Figure 1. Cryo-electron micrograph of the inwardly open conformation of PDR5 (ADP-PDR5). (Harris, A et al., 2021)
Lifeasible, a highly regarded plant biology company, is dedicated to analyzing the plant PDR gene families at the genomic level. We provide efficient, accurate, and customizable bioinformatics analysis solutions for plant multidirectional resistance genes based on PDR gene sequences from multiple databases such as TAIR, NCBI, Benthgenome, and sol genomics.
We will extract the PDR gene and protein sequences of the plants to be analyzed and their ancestral species and important relatives from relevant databases, and use bioinformatics methods to analyze and predict their structure, function, and other characteristics, to provide a scientific basis and theoretical foundation for subsequent work on gene function research, gene editing, and molecular breeding for disease resistance.
Relevant PDR genes will be downloaded from several databases, such as Arabidopsis database TAIR, NCBI, Benthgenome, and solgenomics by BLAST comparison.
The genes downloaded in the previous step are translated into PDR protein sequences using the translation tool on the ExPASY website. Then the physicochemical properties of these proteins are analyzed by the protein parameter analysis tools ProtParam and protscale.
We typically use tools such as TMHMMM, pepwheel, phyre2, and SWISS MODEL to predict the transmembrane, secondary, and tertiary structures of PDR proteins; and SUBA4 to analyze the localization of related proteins in cells online.
CoGe Blast will be used to analyze the similar sequences of PDR genes on each chromosome of these plant genomes.
We can extract a 3000 bp DNA sequence upstream of the PDR gene to be analyzed from the database, use BDGP to analyze the transcription start site of the gene, and use tools such as PlantCare and PLACE to predict the cis-elements of the promoter region of the PDR gene, and then make functional predictions.
The online gRNA design site CRISPR direct is used to design gene editing targets for the PDR gene to be analyzed, using NGG as the PAM sequence, to obtain the CRISPR/Cas9 gene editing targets with the highest score and lowest off-target probability.
We can use the MEME tool to identify the motif and distribution of PDR proteins and Clustal X software to analyze the homologous sequence similarity of PDR proteins. Based on the sequence similarity comparison results, a phylogenetic tree is constructed using MEGA 12.0 with a random sample size of 1000.

Lifeasible provides the most detailed and comprehensive bioinformatics analysis of plant pleiotropic drug resistance genes for our clients. We sincerely hope to enter a collaboration with you. Please feel free to contact us with any questions.
Reference