A New Way to Control Microbial Metabolism Revealed
Microorganisms can be engineered to produce a variety of useful compounds, including plastics, biofuels, and pharmaceuticals. However, in many cases, bacteria need to compete in the metabolic pathways of “self-sustaining” and “synthetic products”.
To help optimize the ability of cells to produce desired compounds and maintain their own growth, chemical engineers at MIT have designed a method that can induce bacteria to switch between different metabolic pathways at different times. These switches are “programmed” into the bacterial genome and are triggered or closed depending on changes in population density without manual intervention.
Kristala Prather, professor of chemistry and the author of the article, said: “We hope this will allow more precise regulation of bacterial metabolism to obtain higher productivity, but at the same time minimize the number of interventions.” This conversion allows researchers to increase the microbial yield of two different products by tenfold.
Christina Dinh, a graduate student at MIT, is the lead author of the paper, which was recently published in
PNAS.
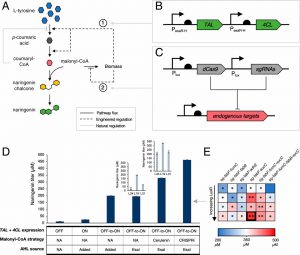
To enable microbes to synthesize compounds that are not normally produced, engineers inserted genes for enzymes involved in metabolic pathways. In some cases, the intermediates produced during these reactions are also part of the metabolic pathways already present in the cell. When cells transfer these intermediates to routes other than the engineering route, the total yield of the final product is reduced.
By using a concept called dynamic metabolic engineering, Prather has previously constructed switches that can help cells maintain a balance between their own metabolic needs and the pathways that produce the desired products. Her idea was to program the cells to automatically switch between pathways without any intervention by personnel operating the reactive fermenter.
The MIT team engineered their
E. coli cells to secrete a quorum-sensing molecule called AHL. When the concentration of AHL reaches a certain level, the cell turns off an enzyme that translocates gluconeic acid into one of the cell's own metabolic pathways. This allows the cells to grow and divide normally until the population is large enough to begin producing large quantities of the desired products.
In the new PNAS paper, Prather and Dinh set out to design multiple switching points into their units, thereby giving them a greater degree of control over the production process. For this purpose, they used two quorum-sensing systems from two different bacteria. They integrated these systems into
E. coli and engineered them to produce a compound called grapefruit, a flavonoid naturally present in citrus fruits with multiple beneficial effects on health.
Using these quorum-sensing systems, the researchers designed two switching points into the cell. A switch was designed to prevent bacteria from transferring the bombesin called malonyl-CoA into the cell's own metabolic pathway. At another transition point, the researchers delayed the production of enzymes in their engineered pathway to avoid the accumulation of excessive amounts that in turn inhibited the synthesis of grapefruit.
The researchers created hundreds of
E. coli variants to execute these two switches at different population densities, thus allowing them to identify which was the most productive. The best performing strains showed a ten-fold increase in bombesin production compared to strains without built-in these control switches.
Reference
Christina V. Dinh, Kristala L. J. Prather. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proceedings of the National Academy of Sciences, 2019; 201911144 DOI: 10.1073/pnas.1911144116 For research or industrial raw materials, not for personal medical use!
 To enable microbes to synthesize compounds that are not normally produced, engineers inserted genes for enzymes involved in metabolic pathways. In some cases, the intermediates produced during these reactions are also part of the metabolic pathways already present in the cell. When cells transfer these intermediates to routes other than the engineering route, the total yield of the final product is reduced.
By using a concept called dynamic metabolic engineering, Prather has previously constructed switches that can help cells maintain a balance between their own metabolic needs and the pathways that produce the desired products. Her idea was to program the cells to automatically switch between pathways without any intervention by personnel operating the reactive fermenter.
The MIT team engineered their E. coli cells to secrete a quorum-sensing molecule called AHL. When the concentration of AHL reaches a certain level, the cell turns off an enzyme that translocates gluconeic acid into one of the cell's own metabolic pathways. This allows the cells to grow and divide normally until the population is large enough to begin producing large quantities of the desired products.
In the new PNAS paper, Prather and Dinh set out to design multiple switching points into their units, thereby giving them a greater degree of control over the production process. For this purpose, they used two quorum-sensing systems from two different bacteria. They integrated these systems into E. coli and engineered them to produce a compound called grapefruit, a flavonoid naturally present in citrus fruits with multiple beneficial effects on health.
Using these quorum-sensing systems, the researchers designed two switching points into the cell. A switch was designed to prevent bacteria from transferring the bombesin called malonyl-CoA into the cell's own metabolic pathway. At another transition point, the researchers delayed the production of enzymes in their engineered pathway to avoid the accumulation of excessive amounts that in turn inhibited the synthesis of grapefruit.
The researchers created hundreds of E. coli variants to execute these two switches at different population densities, thus allowing them to identify which was the most productive. The best performing strains showed a ten-fold increase in bombesin production compared to strains without built-in these control switches.
Reference
Christina V. Dinh, Kristala L. J. Prather. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proceedings of the National Academy of Sciences, 2019; 201911144 DOI: 10.1073/pnas.1911144116
To enable microbes to synthesize compounds that are not normally produced, engineers inserted genes for enzymes involved in metabolic pathways. In some cases, the intermediates produced during these reactions are also part of the metabolic pathways already present in the cell. When cells transfer these intermediates to routes other than the engineering route, the total yield of the final product is reduced.
By using a concept called dynamic metabolic engineering, Prather has previously constructed switches that can help cells maintain a balance between their own metabolic needs and the pathways that produce the desired products. Her idea was to program the cells to automatically switch between pathways without any intervention by personnel operating the reactive fermenter.
The MIT team engineered their E. coli cells to secrete a quorum-sensing molecule called AHL. When the concentration of AHL reaches a certain level, the cell turns off an enzyme that translocates gluconeic acid into one of the cell's own metabolic pathways. This allows the cells to grow and divide normally until the population is large enough to begin producing large quantities of the desired products.
In the new PNAS paper, Prather and Dinh set out to design multiple switching points into their units, thereby giving them a greater degree of control over the production process. For this purpose, they used two quorum-sensing systems from two different bacteria. They integrated these systems into E. coli and engineered them to produce a compound called grapefruit, a flavonoid naturally present in citrus fruits with multiple beneficial effects on health.
Using these quorum-sensing systems, the researchers designed two switching points into the cell. A switch was designed to prevent bacteria from transferring the bombesin called malonyl-CoA into the cell's own metabolic pathway. At another transition point, the researchers delayed the production of enzymes in their engineered pathway to avoid the accumulation of excessive amounts that in turn inhibited the synthesis of grapefruit.
The researchers created hundreds of E. coli variants to execute these two switches at different population densities, thus allowing them to identify which was the most productive. The best performing strains showed a ten-fold increase in bombesin production compared to strains without built-in these control switches.
Reference
Christina V. Dinh, Kristala L. J. Prather. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proceedings of the National Academy of Sciences, 2019; 201911144 DOI: 10.1073/pnas.1911144116