Ancient farmers brought newly domesticated crops into new environments, subjecting them to strong natural and artificial selection pressures, and the survival of plants under such selection pressures was the rapid adaptive evolution of crops. These early crop varieties had genetic and phenotypic diversity, providing sufficient genetic resources to promote environmental adaptation, and eventually formed new varieties adapted to the local area, which was the backbone of early human civilization.
The domestication history of barley (Hordeum vulgare) is a typical example. Barley is an annual plant that was domesticated more than 10,000 years ago and brought from the Fertile Crescent to Europe, Asia and North Africa, becoming the main source of nutrition for humans and livestock for thousands of years. Although the genetic history of barley populations has been partially revealed through germplasm collection and research, the long generation time makes it difficult to study important genetic nodes.
On July 12, 2024, Daniel Koenig's research group at the University of California, Riverside, published a research paper titled "Natural selection drives emergent genetic homogeneity in a century-scale experiment with barley" in Science. Through a long-term experiment, they studied the process of local adaptation of barley over the past century.
Composite cross II (CCII) began in 1929 and aims to adapt genetically diverse populations to environmental conditions in Davis, California, USA. The authors selected 28 varieties that can represent the ecological, phenotypic and geographical diversity of barley and hybridized all combinations, producing more than 20,000 recombinant F2 offspring. This offspring was planted in proportion, and the offspring were allowed to compete freely with minimal human intervention, and the harvested seeds continued to be experimented.
The authors sequenced 28 parents and found that the distribution of SNPs in the parents was correlated with the allele frequency of global genetic variation. Subsequently, the authors selected individuals in each generation of offspring for sequencing and tracked the evolutionary trajectory of each SNP from parent to offspring. The results showed that the genetic diversity of the composite hybrid offspring changed rapidly throughout the experiment, much faster than the rate of variation caused by genetic drift alone. By studying the allele frequencies and genetic distances of the whole genome, the authors found that the genetic diversity was greatly reduced in the F58 generation, indicating that only a few genetic lineages survived to the later stages of the composite hybrid offspring. By Genotyping-by-Sequencing (GBS) of 878 samples from the hybrid parents (F0) and five generations of F18, F28, F50, and F58, and hierarchical clustering of the lineages according to genetic distance, only 261 recombinant selfing lineages were found, among which the recombinant selfing lineage with a very high frequency was named Lin1. As the number of selfing generations increased, the abundance of Lin1 increased sharply, and the genetic diversity of the population also decreased.
To identify the target genes for directional selection of CCII during rapid adaptation, the authors calculated the probability of allele frequency changes in a set of neutral evolution simulations, and after correcting the results, 58 genomic regions were identified. One of the regions contains genes that regulate the flowering time of barley. Based on the above findings, the authors explored how the genetic changes of CCII are transformed into adaptive phenotypic changes by studying the evolution of flowering time genes. By observing the flowering time of parents and different offspring, the authors found that although the median flowering time changed slightly, the outliers of flowering time were decreasing overall, the flowering time was positively correlated with plant height and reproductive capacity, and Lin1 showed a flowering time similar to that of the parents. These results prove that the flowering time selection of CCII is to promote the population to develop towards an intermediate phenotype by eliminating phenotypic extremes. Through allele frequency and genome-wide association analysis (GWAS), it was found that the flowering time of CCII is regulated in two steps: the selection of Ppd-H1, HvCEN and possibly other loci ensures that barley completes its life cycle before the onset of the dry season; the selection of functional Vrn-H2 eliminates premature flowering in winter or early spring.
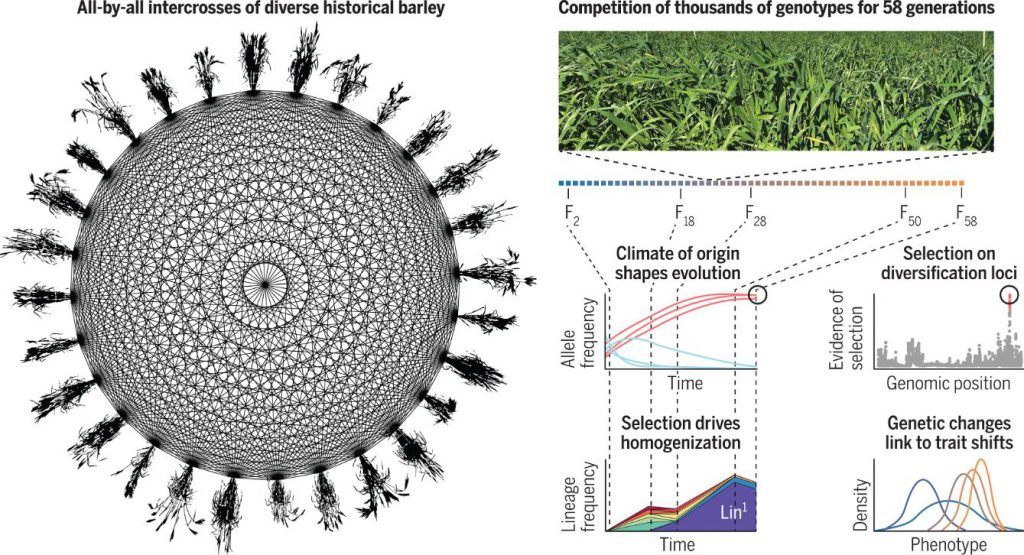
Figure 1. Rapid adaptation in CCII. (Landis, et al., 2024)
In summary, the authors used a composite hybrid population and analyzed the sequencing results of its decades-long selfing population to describe the evolutionary trajectories of millions of genetic variants, explore the sources of alleles favored by natural selection, find target sites of unusually strong selection, and link genetic changes to phenotypic changes over time. The authors found that natural selection dominated the evolution of CCII, the amplitude of natural selection was larger than generally assumed, and the loss of genome-wide diversity was caused by linked selection. Therefore, even with extremely high basal diversity, populations of self-pollinating plants may be doomed to extinction through selection for genetic homogeneity. Composite hybridization makes it possible to explore long-term stabilizing selection mechanisms, helping people to better understand the trade-offs between local adaptation, plant competitiveness and crop yield, helping to develop more resilient crop varieties and improve the understanding of plant evolution.