Rice is the main staple food for more than half of the world's population. Herbicides are the main weed control strategy in large-scale rice cultivation. However, long-term use of herbicides often leads to the evolution of resistant weeds. Additionally, applying herbicides to weeds similar to rice, such as barnyard grass, can have significant negative agricultural impacts. Therefore, discovering and engineering new herbicide resistance genes has become an urgent challenge in Molecular Breeding research.
On August 12, 2024, Guang-Fu Yang and Hong-Yan Lin of Central China Normal University jointly published a research paper titled "An artificially evolved gene for herbicide-resistant rice breeding" in PNAS. The study focused on 4-hydroxyphenylpyruvate dioxygenase Inhibitor Sensitive 1-Like (HSL) protein, which is widely present in higher plants and exhibits weak Catalytic Activity against a variety of β-triketone herbicides (β-THs).
By resolving the crystal structure of the maize HSL1A complex with β-THs, the authors identified four key herbicide-binding residues and explained the reason why HSL1A has weak activity against herbicides. Using artificial evolution methods, the authors developed a series of rice HSL1 mutants targeting these four residues. These mutants were then systematically evaluated, ultimately determining that the M10 variant was the most effective in modifying β-THs. These mutants also reveal the substrate-bound initial active conformation in HSL1. In addition, Overexpression of M10 in rice significantly improved Resistance to β-THs, resulting in a 32-fold increase in resistance to methylbenzoquinone. In summary, the artificially evolved M10 gene shows great potential in developing herbicide-resistant crops.
In 2019, Yuzuru Tozawa et al. discovered a new rice (Oryza sativa L.) gene family, HIS1 (4-hydroxyphenylpyruvate dioxygenase (HPPD) inhibitor sensitive 1) and its homologous gene HIS1-like (HSL). HIS1 encodes a Fe(II)/2-oxoglutarate (2-OG)-dependent oxygenase that detoxifies β-THs through hydroxylation/oxidation. It makes some rice varieties resistant to commercial β-THs such as benzobicyclocopper, sulfonone, benomyl, etc. However, nonfunctional his1 allelic variants were observed in indica rice subspecies, which are susceptible to benzobicyclocopper due to a 28-bp deletion in the HIS1 gene. In contrast, HSL proteins have limited catalytic detoxification effects on β-THs, but HSLs are widely distributed in japonica and indica rice varieties as well as in wheat (Triticum aestivum), corn (Zea mays), and sorghum (Sorghum bicolor). Therefore, Directed Evolution can be used to enhance the detoxification ability of HSLs against HPPD inhibitor herbicides, providing a promising alternative for breeding herbicide-resistant crops.
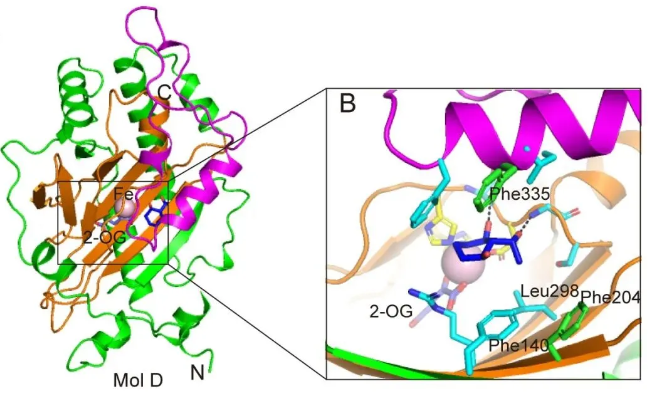
Figure 1. Structure-based artificial evolution strategies. (Dong, et al., 2024)
In this study, the authors report the crystal structure of ZmHSL1A in the apo- and β-TH-bound state and identify four key herbicide-binding residues (F140, L204, F298, and I335 in OsHSL1). The binding mode of β-THs in ZmHSL1A clearly reveals that β-THs are locked in a unique direction, causing the modification site to be far away from the catalytic center, thus explaining the weak activity of ZmHSL1A towards β-THs. Furthermore, to investigate the importance of these four key residues, the authors used an artificial evolution strategy to create a series of OsHSL1 mutants (M1-M10) with single, double, triple, and quadruple mutations. The mutant library was screened using four representative β-THs and a pyrazole herbicide. The M10 mutant (F140H/L204F/F298L/I335F) showed significantly enhanced catalytic activity towards β-THs compared with OsHSL1 wild-type (WT). However, compared to the mutants, only OsHSL1-WT was able to modify topramezone. From these mutants, the authors also report the crystal structure of a functional OsHSL1 mutant bound to a herbicide substrate, revealing the herbicide-bound initial active conformation in OsHSL1. In addition, transgenic expression of the M10 mutant in rice significantly improved resistance to triketone herbicides, resulting in a 32-fold increase in resistance to methylbenzoquinone. This mutant has significant potential in developing herbicide-resistant crops.