As one of the most important economic crops in the world, cotton's genetic improvement and molecular breeding have always been important research directions in the field of plant biology. In recent years, with the rapid development of multi-omics technologies such as single-cell omics, spatial transcriptomics, and spatial metabolomics, scientists have a deeper understanding of the core mechanisms in cotton cell fate transformation, embryonic development, and fiber development. These studies not only provide important theoretical support for revealing the key regulatory networks in the complex development process of cotton, but also open up new ways to improve cotton fiber quality and yield. This article combines two latest research results to systematically explain the key regulatory networks of cotton somatic embryogenesis and fiber initiation development, presenting readers with cutting-edge progress and future directions in the field of cotton research.
Cotton is an important economic crop. Establishing an efficient cotton genetic transformation system is of great significance for accelerating cotton genetic engineering improvement. The cotton transformation system mainly relies on cotton somatic embryogenesis and cotton embryo growth point stem cells. Both transformation systems involve the use of pluripotent stem cells. Therefore, analyzing the acquisition and development of pluripotent stem cells is of great significance for optimizing the cotton genetic transformation system. Early studies mainly focused on the changes in tissue cell morphology during somatic embryogenesis and the analysis of the mechanism of action of somatic embryogenesis marker genes. There is a lack of comprehensive and systematic research on the acquisition of stem cell pluripotency and the somatic embryo development regulatory network during somatic embryogenesis from the single cell level.
On January 20, 2025, Fuguang Li's team from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences used single-cell omics, spatial transcriptomics and spatial metabolomics for joint analysis, and analyzed the regulatory network of somatic pluripotency acquisition and somatic embryo development during cotton somatic embryogenesis from the single-cell level, and identified the key genes that regulate somatic embryo development, providing new insights into cotton cell fate transition, cell fate remodeling and zygotic embryo development, which is of great significance for improving cotton genetic transformation systems and revealing the regulatory network of zygotic embryo development. The relevant research results were published in Nature Communications under the title "Spatiotemporal transcriptome and metabolome landscapes of cotton somatic embryos".
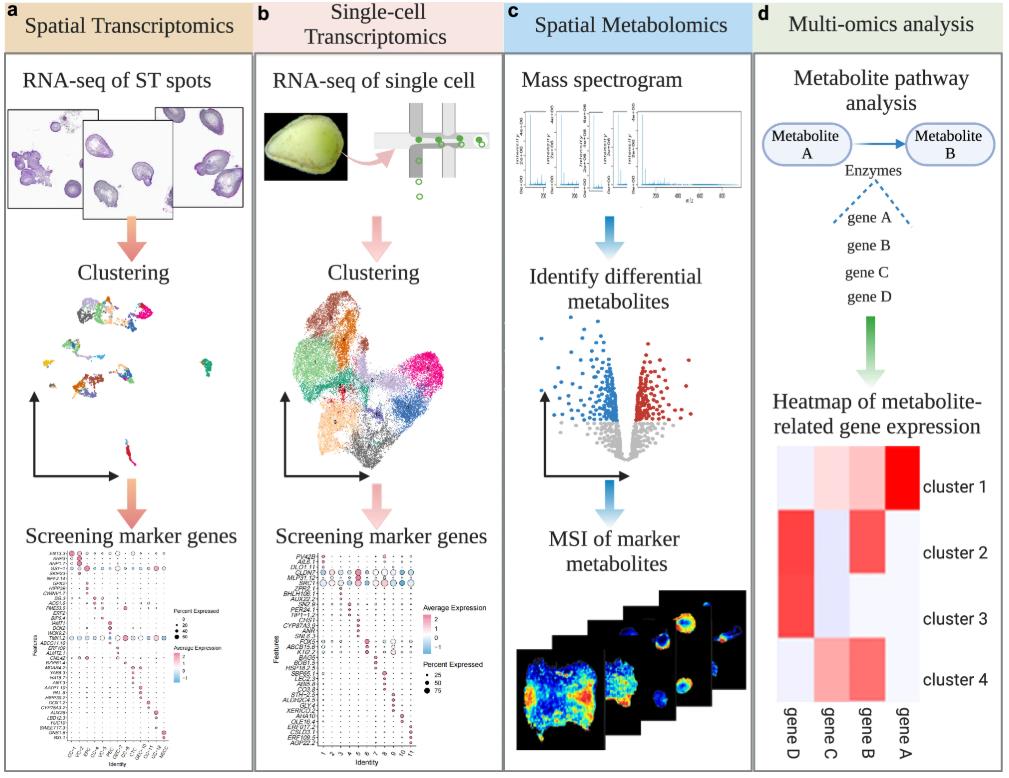
Figure 1. A molecular approach for the development of SE in cotton. (Ge, et al., 2025)
This study used Zhongmiansuo24 (ZM24), a cultivated upland cotton variety with high differentiation efficiency, as the research material. The spatial transcriptional profile and spatial metabolic profile of callus, globular embryos, torpedo embryos, and cotyledon embryos of ZM24 were mapped using single-cell omics, spatial transcriptomics, and spatial metabolomics, and a multi-level dynamic regulatory network of cotton somatic embryogenesis was constructed. A number of key genes that regulate cell pluripotency and somatic embryo development were identified, confirming that the AATP1 and DOX2 genes play a key role in regulating the acquisition of pluripotency and somatic embryo development. Due to the difficulty in sampling cotton zygotic embryos, there has been little progress in the analysis of their developmental regulatory network. The development of cotton somatic embryos is highly similar to that of cotton zygotic embryos. The analysis of the cotton somatic embryo development regulatory network provides theoretical support for analyzing the regulatory mechanism of cotton zygotic embryo development and improving cotton seed development. In addition, to visualize the spatiotemporal expression patterns of key genes regulating somatic embryogenesis, single-cell omics data and spatial transcriptome data of cotton somatic embryogenesis have been shared.
Cotton is the main raw material for the textile industry. Cotton fiber develops from a single cell on the surface of the seed epidermis. In the early stage of seed development, about 25% of the ovule epidermal cells differentiate into cotton fibers and go through four different stages, namely the initiation stage (2 to 5 days after flowering), elongation and primary wall formation stage (3 to 20 days after flowering), secondary wall thickening stage (16 to 40 days after flowering) and fiber maturation stage (40 to 50 days after flowering). The first two stages determine the number and length of fibers produced by each seed, which will directly affect the final cotton fiber yield. The last two stages are related to cell wall thickening, which determines the strength and fineness of the fiber, which will directly affect the final cotton fiber quality. However, the regulatory mechanisms that determine these developmental stages are still largely unknown. Therefore, analyzing the mechanisms that determine the fate of cell development and regulate the initiation and elongation of cotton fibers will provide new ways to improve cotton fiber yield and quality.
On January 20, 2025, Xiongfeng Ma's team from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences successfully constructed the first cotton fiber initiation development profile that combines single-cell transcriptome, spatial transcriptome and spatial metabolome. This profile can identify the expression patterns of key genes and their relationship with metabolic pathways, deeply analyze the core regulatory mechanisms in the fiber development process, and provide new ideas for further improving fiber quality and increasing fiber yield, and provide important theoretical support for high-quality cotton molecular breeding. The relevant research results were published in Nature Communications under the title "The spatiotemporal transcriptome and metabolome landscapes of cotton fiber during initiation and early development".
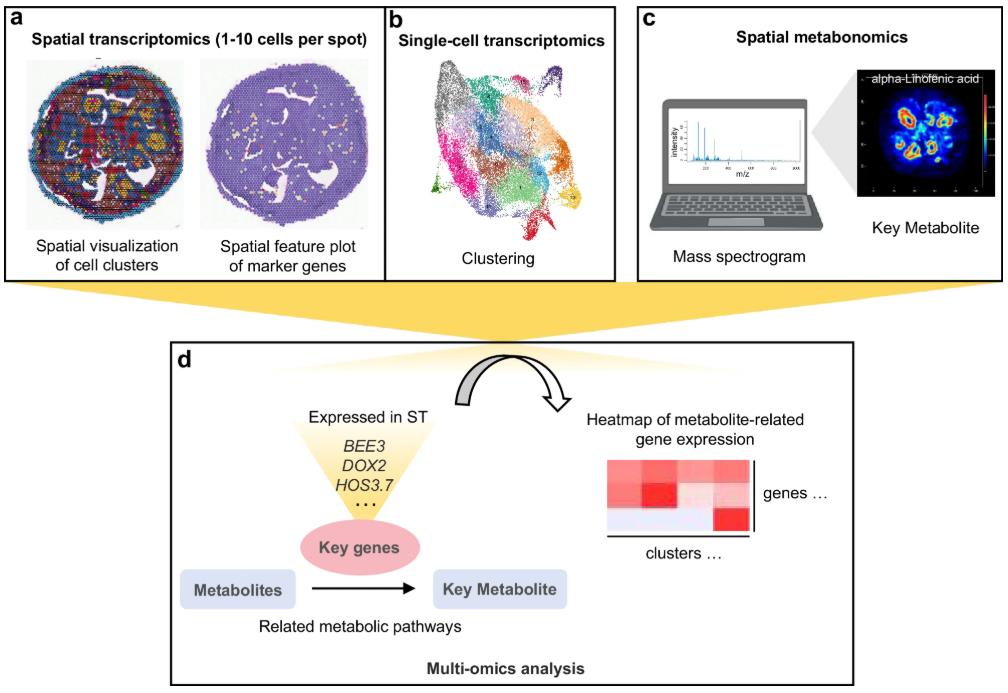
Figure 2. A molecular approach towards fiber development atlas in cotton. (Sun, et al., 2025)
The study used the standard line of upland cotton TM-1 as the research material, mapped the single-cell transcriptome profile, spatial transcriptome profile and spatial metabolome profile of cotton fiber early development, and constructed a multi-level dynamic regulatory network for cotton fiber initiation development, which can not only capture the gene expression pattern during fiber cell development with high resolution, but also reveal metabolic changes closely related to gene expression, filling the current technical gap in fiber cell development research. This study further identified a number of key genes that regulate fiber initiation and development through multi-omics joint analysis, and selected some of these genes for functional verification. It was found that the BEE3 gene plays a key role in regulating the process of fiber initiation and development. Overexpression of this gene can significantly promote the development of ovule epidermal cells into fiber cells. The functional mechanism of this gene is currently being genetically analyzed in the high-lint material Zhongmian 113, and excellent allelic variations are being discovered, in order to develop molecular markers with breeding value for the cultivation of high-lint cotton varieties. The research results comprehensively analyzed the dynamic regulatory network in the process of fiber initiation and development at the single-cell level, providing important data resources for the study of cotton fiber development and theoretical support for cotton precision breeding. The single-cell omics data and spatial transcriptome data in the process of cotton fiber initiation and development have been shared.
These two studies started from the two key biological processes of somatic embryogenesis and fiber initiation and development, respectively, and used multi-omics technology to map the dynamic regulatory network of cotton development with high precision. These research results not only systematically analyzed the molecular regulatory mechanism in the fate transformation and development of cotton cells, but also filled the technical gap in cotton genetic transformation and fiber quality research.
In the future, with the further development and integration of multi-omics technology, these research methods can also be widely used in the analysis of other complex biological processes of cotton. At the same time, through functional verification and molecular marker development, these research results will provide important support for cotton molecular breeding, help cultivate new cotton varieties with excellent fiber quality, high yield and stress resistance, and contribute to the sustainable development of the global cotton industry.