Since the Arabidopsis genome has been sequenced, when studying the promoter of a certain gene, primers can be designed based on the known gene sequence and the promoter sequence of the gene can be directly cloned. However, for plant materials such as Rosa chinensis and Lolium, since there is no known genome sequence, genome walking needs to be used when cloning the promoter of a certain gene.
Commonly used chromosome walking methods include inverse PCR, panhandle PCR, ligation-mediated PCR, thermal asymmetric interlaced PCR (TAIL-PCR), and single oligonucleotide nested PCR (SON PCR).
The basic principle is to design three specific primers (SP Primers) in the same direction and with high annealing temperature and a degenerate primer with lower annealing temperature based on the known DNA sequence to perform a thermal asymmetric PCR reaction.
1. Design of specific primers
Based on the verified known sequences, three specific primers were designed according to the specific primer design principles, named SP1, SP2, and SP3 respectively.
2. First round of PCR reaction
Take an appropriate amount of genomic DNA as the template, use AP1 Primer as the upstream primer, and SP1 Primer as the downstream primer. Add each component according to the reaction system in Table 1, mix and centrifuge briefly. Set the PCR machine according to the PCR1 reaction program. Part of the obtained product was diluted 1 to 1000 times (depending on the product concentration) and used as a template for the second round of PCR reaction.
Table 1. Reaction System for The First Round of PCR
| Component | Genomic DNA | dNTP | 10X LA Buffer | LA Taq | AP1 | SP1 | ddH2O |
| Amount | 1-10 ng | 2 µL | 5 µL | 0.5 µL | 1 µL | 1 µL | Make up to 50 µL |
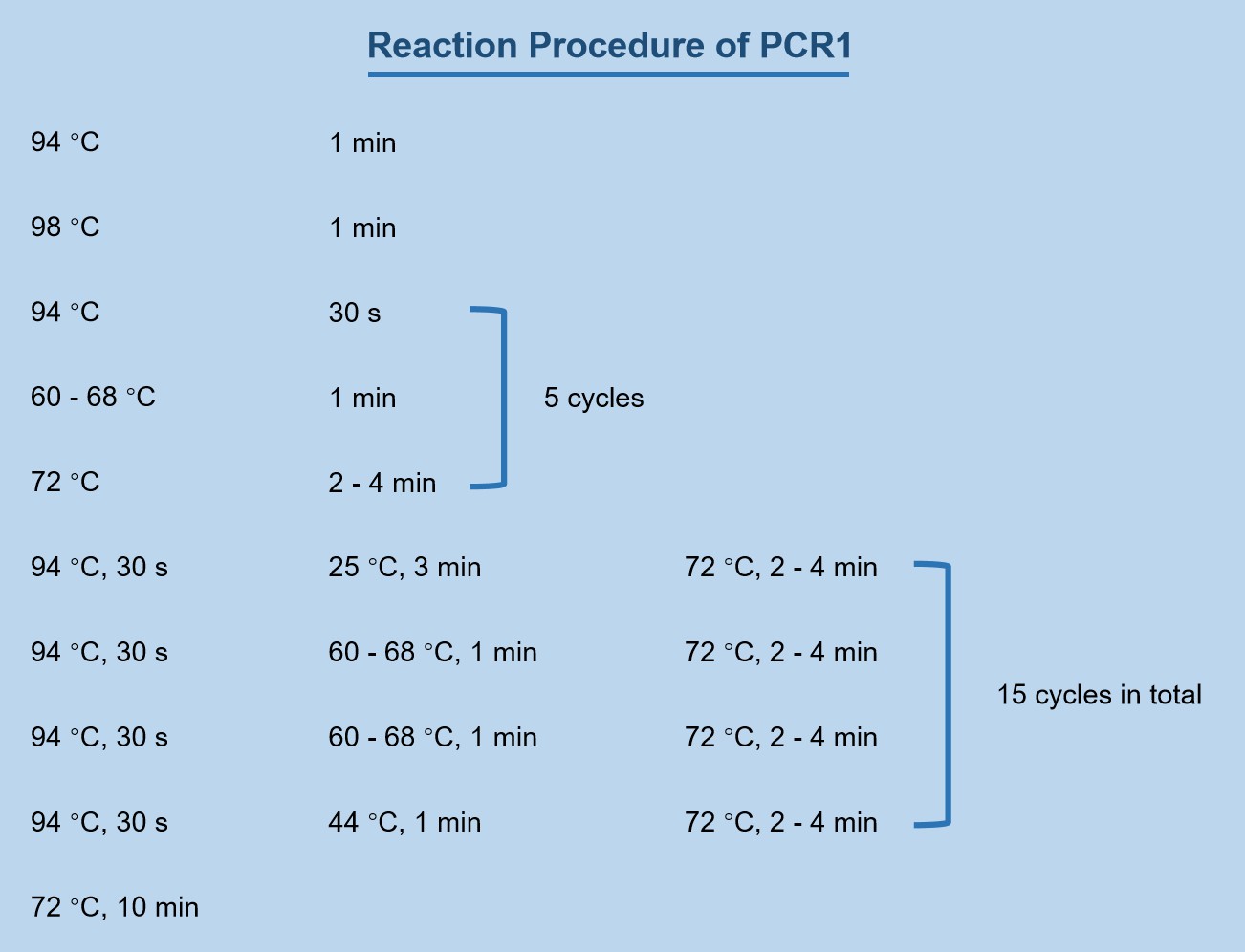 Figure 1. Reaction Procedure of PCR1
Figure 1. Reaction Procedure of PCR1
3. Second round of PCR reaction
Take 1 µL of the dilution of the first round reaction product as the template for the second round of PCR reaction, use AP1 Primer as the upstream primer and SP2 Primer as the downstream primer to perform the second round of PCR reaction. Add each component according to the reaction system in Table 2, mix and centrifuge briefly. Set the PCR machine according to the reaction program of PCR2. Part of the obtained product was diluted 1 to 1000 times (depending on the product concentration) and used as a template for the third round of PCR reaction.
Table 2. Reaction System for The Second Round of PCR
| Component | Dilutions of First Round Product | dNTP | 10X LA Buffer | LA Taq | AP1 | SP2 | ddH2O |
| Volume | 1 µL | 2 µL | 5 µL | 0.5 µL | 1 µL | 1 µL | Make up to 50 µL |
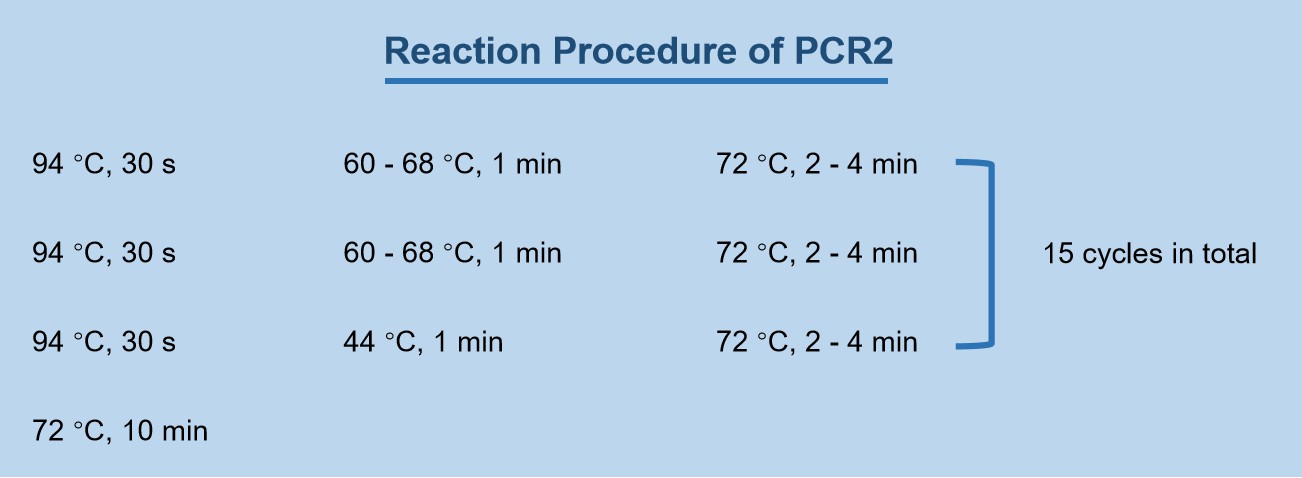 Figure 2. Reaction Procedure of PCR2
Figure 2. Reaction Procedure of PCR2
4. The third round of PCR reaction
Take 1 µL of the dilution of the second round reaction product as the template for the third round of PCR reaction, use AP1 Primer as the upstream primer and SP3 Primer as the downstream primer to perform the third round of PCR reaction. Add each component according to the reaction system in Table 3, mix and centrifuge briefly. The reaction procedure of PCR3 is the same as that of PCR2.
Table 3. Reaction System for The Third Round of PCR
| Component | Dilutions of The Second Round Product | dNTP | 10X LA Buffer | LA Taq | AP1 | SP3 | ddH2O |
| Volume | 1 µL | 2 µL | 5 µL | 0.5 µL | 1 µL | 1 µL | Make up to 50 µL |
5. Result verification
Take 5 µL of each of the first, second, and third round PCR reaction solutions and conduct electrophoresis on a 1% agarose gel. Clear electrophoresis bands were recovered by cutting the gel, and the PCR products were verified by DNA sequencing using SP3 Primer as primer.
The annealing temperature in each round of reaction is determined based on the Tm of the specific primer.