Individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) is a refinement of CLIP that can achieve a resolution to one nucleotide of RBP binding sites. As a global leading biotechnology company, Lifeasible offers customers professional and first class iCLIP analytical services.
iCLIP was developed as an efficient approach to identify cross-linking sites of RNA-protein interactions at a single-nucleotide resolution. This method takes advantage of the fact that reverse transcriptions are truncated at the nucleotides that are crosslinked to peptides and remain modified after proteinase K digestion. As other CLIP methods, iCLIP uses UV light to covalently bind proteins and RNA molecules. The RNA-protein complexes were separated using immunoprecipitation followed by SDS-PAGE and membrane transfer. However, an alternative strategy of adaptor ligation is employed in iCLIP to include the truncated cDNAs in the library. Adapters are added to the 5’ cDNA ends instead of 5’ RNA ends and then cDNAs were cyclized and linearized for PCR and sequencing. The modified cDNA library preparation protocol allows for identification of the cross-linking sites at high resolution (Figure 1).
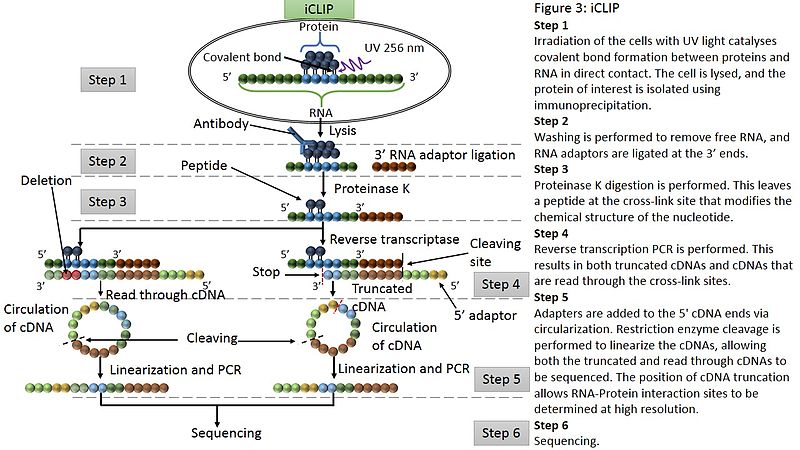 Figure 1. The workflow of an iCLIP assay (Wikipedia)
Figure 1. The workflow of an iCLIP assay (Wikipedia)
We are devoted ourselves to providing customized one-stop services for the iCLIP assays, including sample preparation, Immunoprecipitation, proteinase K treatment, reverse transcription, cDNA library preparation, sequencing, and data analysis and report. We are also dedicated to developing advanced methods to meet customer specific requirements. Our services are featured with high reliability, short turnaround time, and competitive price. Welcome to contact us for further information.