The yeast 2-hybrid (Y2H) assay is a very powerful tool for detecting protein-protein or protein-DNA interactions. Since its discovery in the 1980s, this assay has become one of the most popular approaches for probing physical interactions in vivo. The Y2H assay is not only straightforward to perform, but also highly feasible and can be applied in a broad categories of proteins, including those in plants. Here at Lifeasible, we provide well established platforms for Y2H assays with guaranteed high quality results and rapid turnaround time.
The Y2H Assay
The principle of a traditional Y2H assay is to utilize a reporter gene, whose activation relies on its particular transcription factor (1). Specifically, proteins to be tested, X and Y, are fused with the DNA-binding domain (BD), and the activation domain (AD) of the transcription factor, respectively. The BD-X fusion protein binds to the upstream activator sequence (UAS) the reporter gene’s promoter. Upon interaction between X and Y, the AD will be recruited to the promoter and reconstitute a functional transcription factor, which therefore triggers the subsequent transcription of the reporter gene (Figure 1).
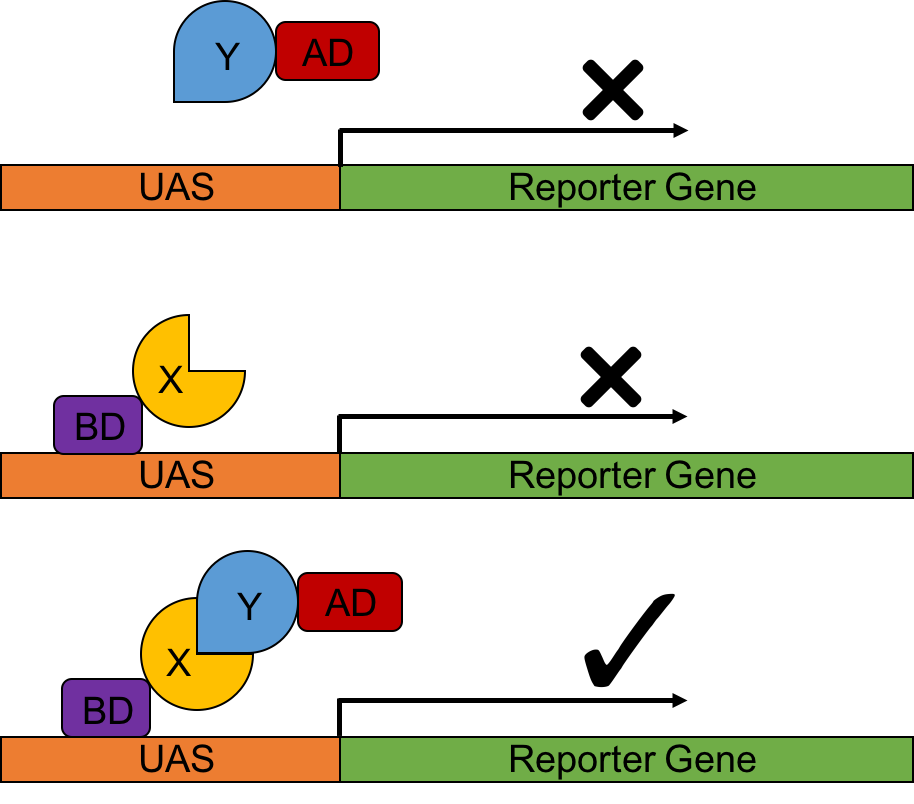 Figure 1. A schematic cartoon of a classical yeast two hybrid system.
Figure 1. A schematic cartoon of a classical yeast two hybrid system.
In order to study the interactions of a broad group of proteins, several variants of the Y2H systems have been developed, adapting to the differential subcellular localizations and biochemical characterizations of different proteins. Lifeasible provides service for multiple variations of the Y2H system to accommodate the diverse needs of different projects.
 Figure 2. A schematic cartoon of the yeast one hybrid system.
Figure 2. A schematic cartoon of the yeast one hybrid system.
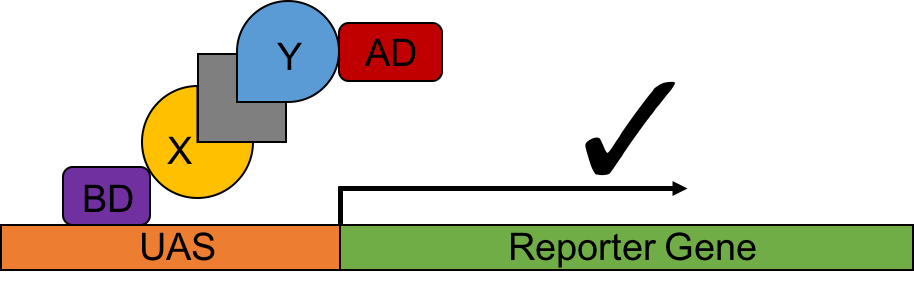 Figure 3. A schematic cartoon of the yeast three hybrid system.
Figure 3. A schematic cartoon of the yeast three hybrid system.
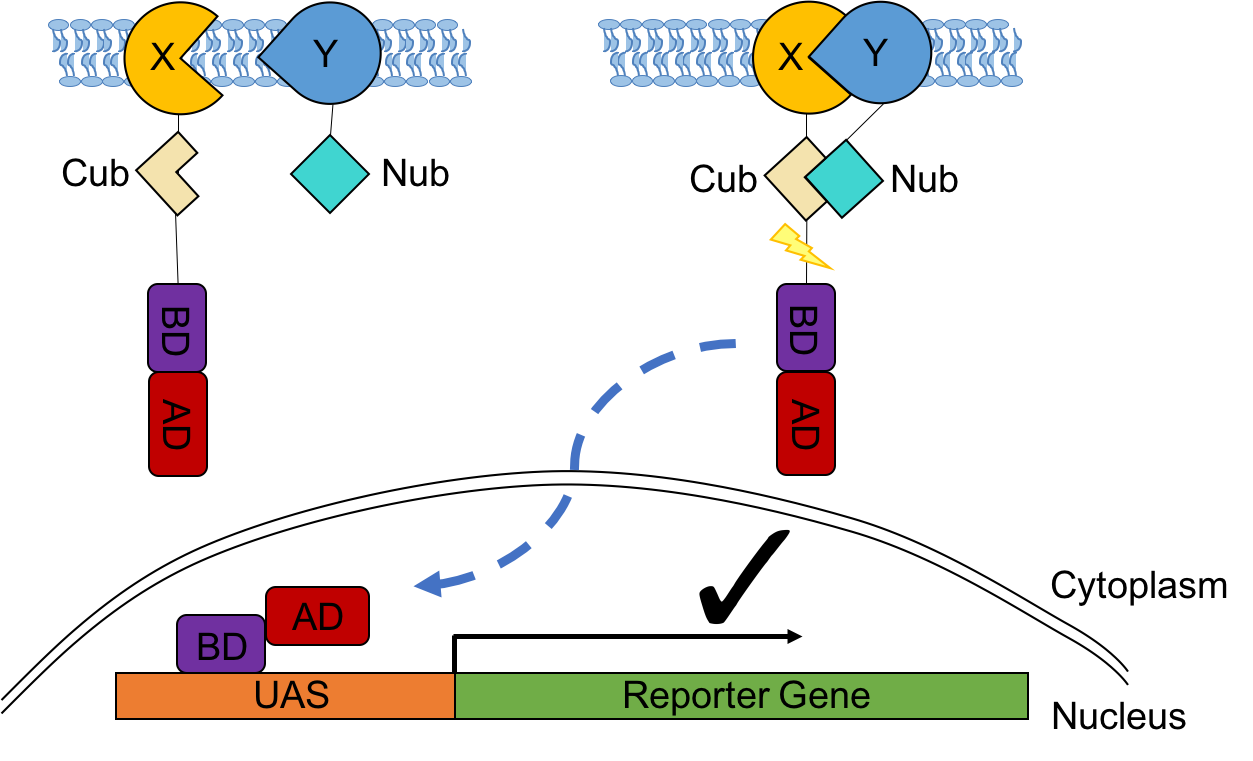 Figure 4. A schematic cartoon of the split-ubiquitin Y2H system.
Figure 4. A schematic cartoon of the split-ubiquitin Y2H system.
Y2H screening
When investigating the function of an unknown protein, it is essential to identify its interactive partners. Aside from examining interactions between noted proteins/DNAs, the Y2H technique has also been widely used as a screening method for the uncovering of protein-protein interactions. Based on the construction of cDNA libraries, Y2H can be easily applied for high throughput screens of protein interactions on a genome-wide base (5). So far, the Y2H screen has been applied for protein functional identifications in the model plant Arabidopsis and multiple crops, including wheat and maize (5, 6).
Advantages of the Y2H screening are:
Reference