BSA is a widely used genetic mapping method to analyze genomic loci affecting a trait of interest. The technology usually mixes individuals with extreme traits in a separated group to build two mixed pools, then the molecular markers closely linked to the target gene can be quickly located by analyzing the frequency difference of alleles between the two mixed pools. BSA overcomes the limitation of obtaining near-isogenic lines in many crops, thus it can save a lot of time. It is a very practical method for gene marker positioning.
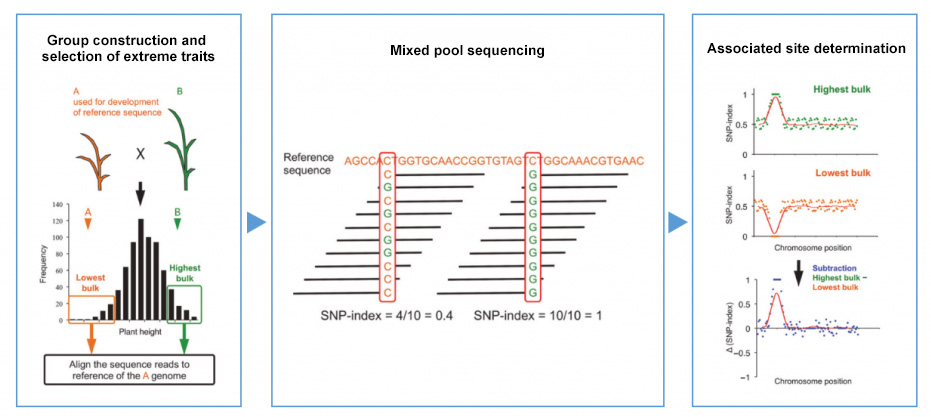 Figure 1. Bulk Segregant Analysis (BSA) Service flowchart
Figure 1. Bulk Segregant Analysis (BSA) Service flowchart
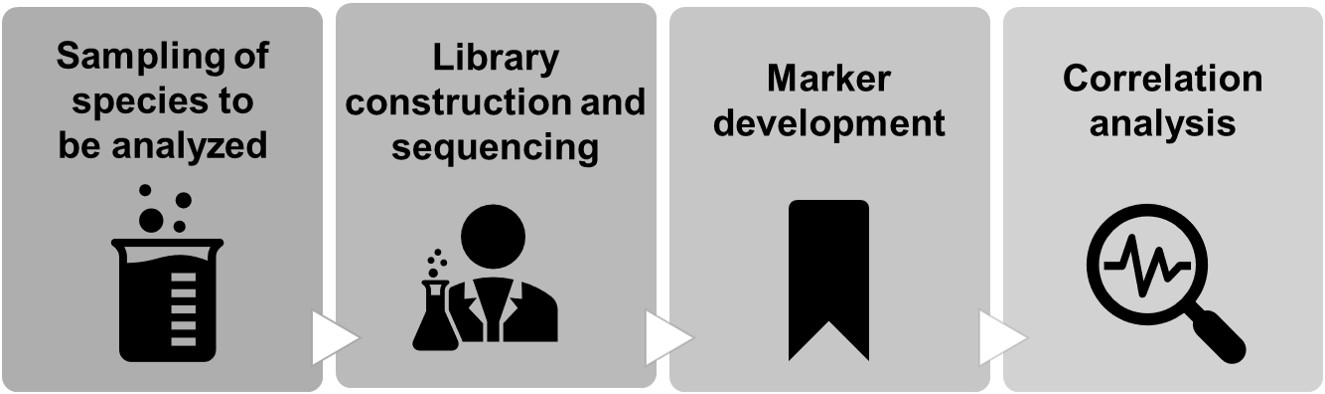
The BSA analysis provided by Lifeasible is fast, accurate, and cost-effective. It is widely used in dozens of traits of many species. At the same time, we will also provide customized analysis solutions for specific groups and traits. The process of a customized analysis solution from sampling scheme design, DNA extraction, library construction and sequencing to analysis:
According to the genome size, complexity, and research purpose of the species, Lifeasible provides our customers with customized analysis strategies, including resequencing-BSA and its mutant analysis form Mutmap, SLAF-BSA and transcriptome-BSA (BSR).
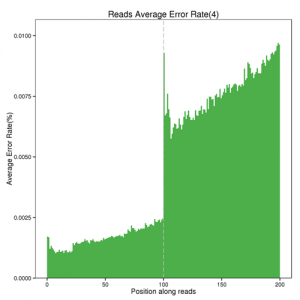
High-quality sequencing data is an important basis for subsequent analysis. After obtaining the raw data for sequencing, we will perform various sequencing evaluation methods such as sequencing data output statistics, base sequencing quality distribution, base type distribution, etc., to ensure the accuracy of the sequencing data.
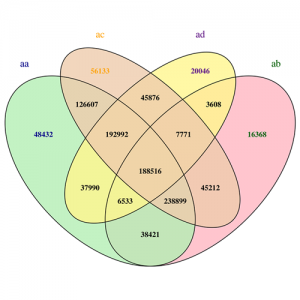
SNP (Single Nucleotide Polymorphism) and InDel (Insertion-Deletion) are the most common genetic markers in the genome, which are characterized by large numbers and rich polymorphisms. The detection of SNP and InDel is mainly realized through GATK and samtools software toolkit, SnpEff software was used for annotation of variant sites. We have rich experience in genetic variation analaysis.
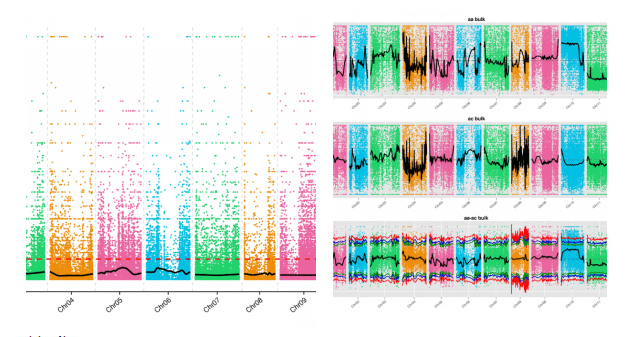
According to the SNP and Indel markers of the parents and the mixed pool, the SNP-Index and ED analysis methods are used to perform the association analysis between the markers and the traits at the same time to obtain the genes related to the target traits.
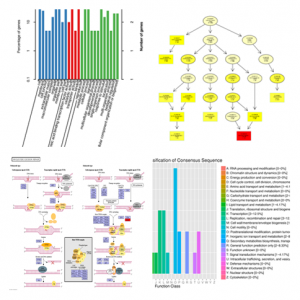
Use BLAST software to perform in-depth annotation of multiple databases (NR, Swiss-Prot, GO, KEGG, COG) for coding genes in the candidate interval. Through detailed annotations, further narrow the associated regions and quickly screen candidate genes.
Lifeasible has successfully completed the positioning of more than 1,000 traits for nearly 300 species. With rich project experience, we can choose the best analysis plan according to our customer’s project needs to ensure accurate analysis results.
For more information or any inquiry requirements, please contact Lifeasible.