For the few species whose genomes have been sequenced (e.g., human, mouse, nematode, rice, Arabidopsis, etc.), the sequences flanking the known sequences of a particular species can be easily found in databases. However, for most organisms, the way to know the DNA sequences flanking a known region without knowing their genome sequence is to use DNA walking techniques. DNA walking is an important molecular biology research technique that can be used to obtain unknown sequences adjacent to known sequences efficiently.

There are more than a dozen PCR-based DNA walking techniques, and Lifeasible can provide you with TAIL-PCR (thermal asymmetric interlaced PCR). It uses specific nested primers, combined with concatenated primers to fish for flanking sequences.
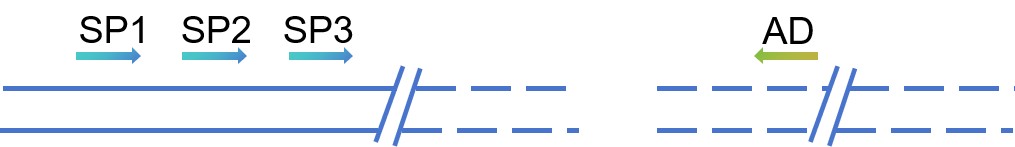
We design three nested specific primers (SP1, SP2, SP3) using known sequences next to the target sequence and use each of them in combination with a short, random, degenerate primer (AD) with a low Tm value. An asymmetric temperature cycle is then designed according to the differences in primer length and specificity, and the specific product is amplified by a graded reaction. TAIL-PCR technology has the advantages of simplicity, speed, high specificity, accurate and reliable reaction products, and good reproducibility.
Lifeasible provides DNA walking technology services to help you identify the order in which specific DNA fragments have been cloned and flanked. Please feel free to contact us to submit your requirements.