Forage grasses are polyploid and occur in various life forms that allow for more than two sets of basic chromosomes. The high incidence of whole genome duplication in forage grasses has prompted breeders to develop new polyploid forage germplasm with higher economic value. Forage grasses are widely exploited as nutrients for animal feed or biofuels. Polyploidy is a widely distributed phenomenon among forage grass species. Induction in diploid species can improve biomass productivity and nutritional properties. Plant breeders have been successful in obtaining polyploid breeding germplasm in a wide range of forage grasses by optimizing methods, concentrations, and timing, especially when colchicine is used. These experimental forage polyploids are valuable tools for understanding gene expression driven by dose-dependent gene expression, altered gene regulation, and epigenetic changes.
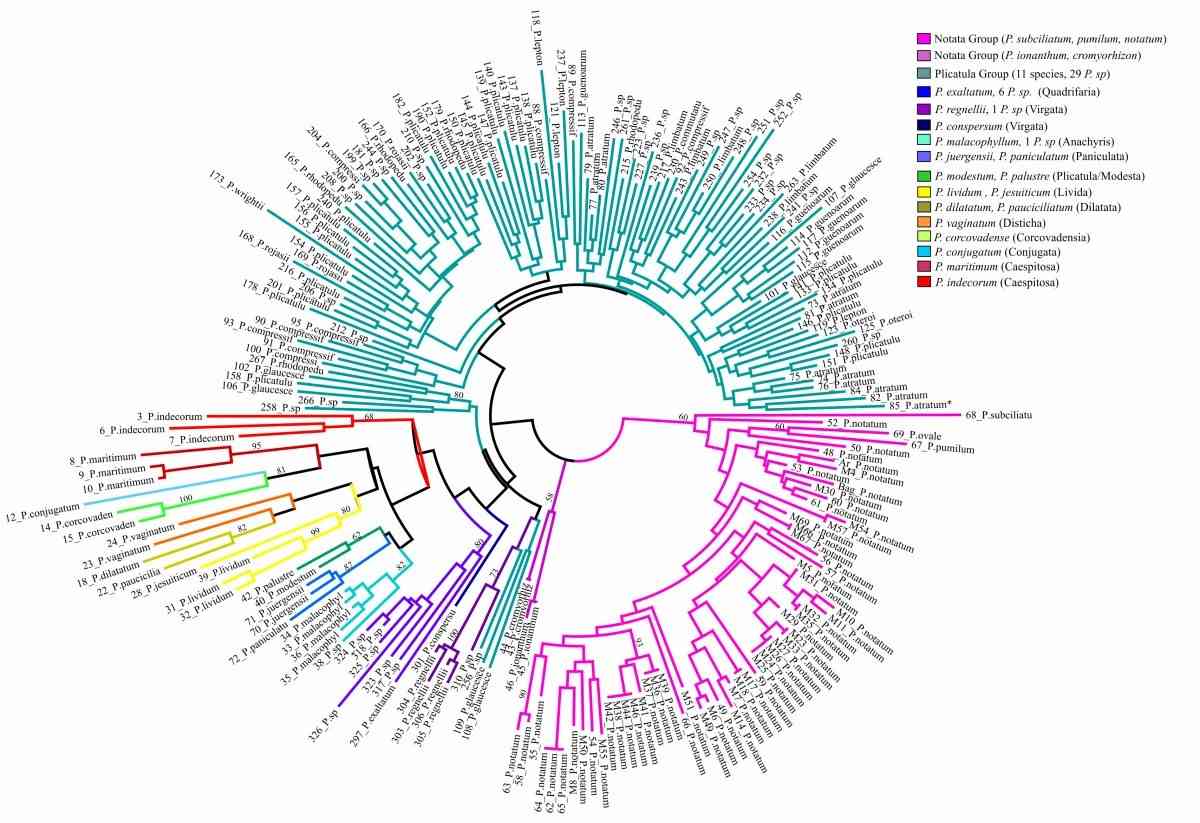 Fig.1. Detailed unrooted neighbor-joining tree based on Dice’s similarity coefficient for the 214 Paspalum accessions. (Cidade F W et al., 2013).
Fig.1. Detailed unrooted neighbor-joining tree based on Dice’s similarity coefficient for the 214 Paspalum accessions. (Cidade F W et al., 2013).
Forage grasses have complex and large polyploid genomes, which makes the application of marker-assisted selection more challenging than in diploid grasses such as rice and maize. As an ideal partner for forage research, Lifeasible offers customized solutions for genetic variation in polyploid forages to assess their genetic differences and classify them as appropriate plants. Our molecular genetics platform is widely used for forage species identification in preliminary germplasm resource assessment and for characterization, conservation, and successful species hybridization. We offer the following methods to produce polyploid forage grasses:
In addition, we offer screening, selection, and periodic evaluation of natural and induced ploidy level stability, depending on customer needs for pure polyploids of different ploidy levels obtained after chromosome doubling. Lifeasible has a powerful ploidy level screening tool flow cytometry to improve the efficiency of forage DNA ploidy level determination and nuclear genome size measurement, enabling the use of quantitative, in one day fast, reliable, and reproducible method to rapidly assess large numbers of individuals in a single day. Our microscopy platform performs chromosome counts by observing mid-term cells to confirm chromosome numbers and detect eventual aneuploidy.
We provide isozymes and DNA-based molecular markers to identify rare alleles at forage specific loci, which are further used to select suitable parental breeding lines, including heterozygosity, phenotypic plasticity, and adaptation to different environments. In addition, we develop transcriptomics and proteomics to analyze the nutritional advantages of polyploids of forage grass species such as alfalfa. Further studies by our expert team on induced polyploid forage grasses may also include the analysis of interference with increased nuclear gene copies for organelle gene expression in mitochondria and plastids.
Induced polyploidy has many positive effects compared to diploidy in various forage species.
We develop multiple methods of inducing forage polyploidy to increase forage biomass production, persistence and regeneration after grazing, and better abiotic stress tolerance. As well as to improve the stability of induced forage polyploids by selecting for balanced chromosome pairing and segregation. Our solutions for genetic variation in polyploid forage grasses are highly regarded by our customers worldwide. For more information or to discuss in detail, please contact us.
Reference