Network Targeting and New Target Prediction
Inquiry
The discovery and elucidation of active substances in herbal medicines and the discovery of targets are central propositions in herbal medicine modernization research. However, obtaining chemical components using analytical chemistry, molecular biology, and biochemistry techniques for experimental validation to find possible targets has the disadvantage of high processing costs, high workload, and slow progress. Lifeasible uses network targeting and new target prediction schemes to construct drug action or related disease networks from different perspectives, build analytical and predictive models, study drug action mechanisms and discover new targets.

From a chemical small molecule perspective
We take a known small molecule (with a known structure and target) as a template to find the target of an unknown small molecule by comparing its structure, based on which we build a network of targets and establish a relationship with the disease. The basic idea is that molecules with similar structures may have the same function, i.e., act on the same targets. We can use the Target Prediction Tool to determine the characteristics of a lead molecule by analyzing the chemical and active space of the molecule to support your work on new target prediction.
- ScaffoldHunter
- ChEMBL
- BindingDB
- Machine learning-based predictive analysis of new drugs in the G-protein coupled receptor family and protein kinase family
- Bayesian models
From a target perspective
We build disease-related networks and use them to obtain structural information (primary sequences or 3D structures) about the target, then carry out high-throughput computer screening to find the active chemical molecule. This approach is more fine-grained and specific than the traditional targeting of an enzyme and avoids off-target effects or complete inhibition of the enzyme's activity. We have developed a predictive target prediction model based on chemogenomics, chemoinformatics, and pharmacology, providing a new tool for the multi-target prediction of drugs.
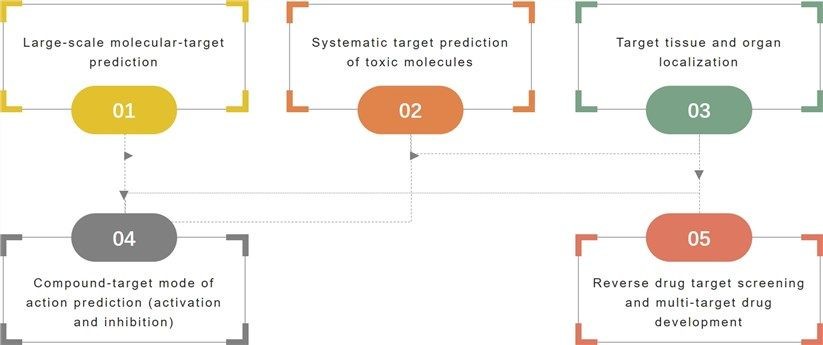
- Systematic targeting. A large-scale molecular network target prediction technique based on chemistry, genomics, pharmacology, and systems analysis techniques.
- Systematic toxicity target prediction. Large-scale toxic-network target prediction techniques based on chemistry, genomics, toxicology, and systems analysis techniques.
- Target tissue localization (TTL). Large-scale target-organ localization techniques based on genomics and microarray analysis techniques.
- Reverse network targeting screening (rNTS). Large-scale target network molecular screening techniques based on chemistry, genomics, pharmacology, and systems analysis techniques.
- Drug mode of action prediction. Large-scale molecular network target mode of action prediction techniques based on chemistry, genomics, pharmacology, and systems analysis techniques.
Applications
The complexity of the herbal system is such that Lifeasible uses high-content, high-throughput omics techniques based on this feature to obtain extensive omics data on the interactions between drugs and experimental models (cells, tissues, etc.) and then uses computational biology to construct drug-target network-disease network models. Please feel free to contact us for an in-depth discussion of the pharmacological mechanisms of herbal medicines.
For research or industrial raw materials, not for personal medical use!
Related Services


