Lifeasible focuses on the role of effector proteins in plant-pathogen interactions and offers services to help study the role of effector proteins. Here, we offer identification and discovery services for effector proteins.
Pathogens produce various effectors for colonizing host plants, including secreted proteins, secondary metabolites, and small non-coding RNAs. Studying these effectors contributes to the understanding of plant disease formation processes and to the development of new strategies to fight pathogenic infections. For example, the study of rust pathogen effectors contributes to wheat resistance breeding (Fig. 1). Among these effectors, effector proteins play an important role in plant-pathogen interactions. Many secreted proteins have been found to be proteases or protease inhibitors. They dismantle host perception machinery, disrupt host defense structures, or degrade/inhibit host-derived proteases. Lifeasible focuses on plant-pathogen interactions and contributes to the study of effector proteins. Here, we offer professional services to help identify and discover effector proteins.
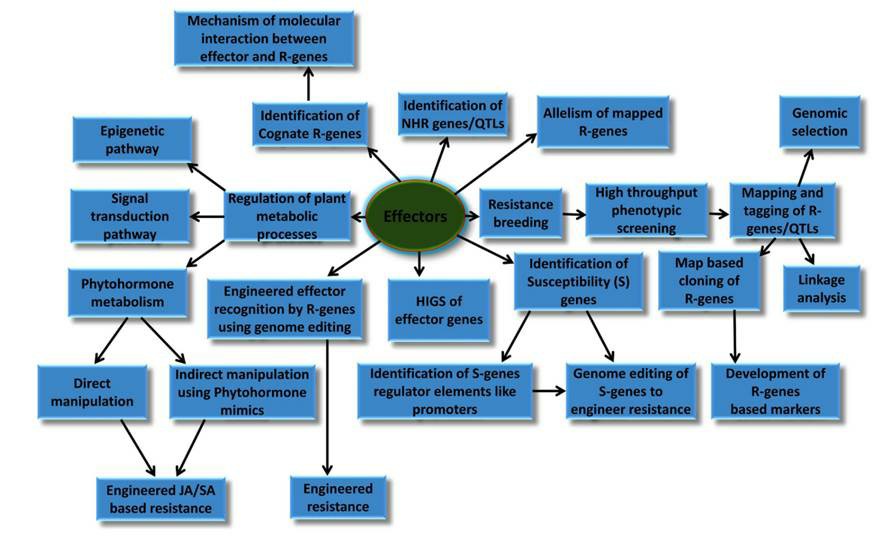 Fig. 1 Potential applications of effectors in wheat resistance breeding and molecular plant biology (Prasad et al., 2019).
Fig. 1 Potential applications of effectors in wheat resistance breeding and molecular plant biology (Prasad et al., 2019).
Lifeasible offers professional services in effector protein identification, and we have extensive experience in bioinformatics-based effector protein discovery. The following are our three main strategies for identifying and discovering effector proteins.
Identification of effector proteins based on secretion properties
The effector proteins are secreted into the host apoplast or cytoplasm to function. We utilize this property for the identification of effector proteins. We can identify them based on secretion-related signal peptides and signature motifs associated with secretion. For example, we can identify effector proteins using the motif RxLR, a conserved translocation motif required for translocation into the host cell.
Identification of effector proteins based on structural properties
The structural properties of proteins are more conserved than the amino acid sequence. We help utilize structural properties to discover effector proteins that are very different in sequence but similar in structure. We have in-depth knowledge of the structural features of a wide range of effector proteins. We can analyze candidate effector proteins using methods such as 2D/3D nuclear magnetic resonance (NMR), cryo-electron microscopy (cryo-EM), and X-ray crystallography. We can then identify them based on the structural homology to characteristic protein scaffolds or structural domains of the identified effector proteins. For example, some effector proteins contain a C-terminus 3-α-helix fold or a β-sandwich fold, and we help identify effector proteins with these characteristics.
Deeper mining of genomic data
The development of second-generation sequencing and third-generation sequencing has facilitated the collection of genomic information. We make full use of the genomic information of pathogens to discover effector proteins. We can use taxonomic information and genomic features of potential candidates to discover effector proteins. Highly variable regions of the genome contribute to the adaptive colonization of pathogens, and we help identify potential effector proteins based on the variability of the genomic region and the proximity to transposable elements (TEs). In addition, highly conserved genetic regions in the pathogen classification of pathogens that infect the same host contribute to the colonization of these pathogens. We help analyze these conserved regions to identify potential effector proteins. Furthermore, we also help to identify effector proteins through gene annotation and identify genes in the genome that are co-regulated in the disease process.
Lifeasible provides professional effector protein identification services, and we master various effector protein identification and discovery strategies. Please contact us to discuss your project.
References