Most eukaryotic expressed proteins need to undergo a series of post-translational processing and modifications to form the final complex functional executor, making post-translational modification of proteins an important aspect of proteomics research. Currently, the main protein modifications that have been studied on a large scale include Phosphorylation, Acetylation, Glycosylation, Ubiquitination, etc. Among them, Phosphorylation, Acetylation, Glycosylation, Ubiquitination, etc. have been studied on a large scale. Phosphorylation is the most common and important post-translational modification of proteins, occurring on serines, threonines and tyrosines, and is regulated by two enzyme families with opposite roles - protein kinases and phosphatases.
Protein phosphorylation modification analysis techniques include protein phosphorylation identification (purified protein phosphorylation identification/phosphorylated protein full spectrum identification) and protein phosphorylation quantification. Lifeasible provides phosphorylation modification analysis services for insect proteins to quantify the phosphorylation of insect proteins.
| Analysis content | Detail |
| Protein phosphorylation modification labelling quantification |
|
| Quantification of protein phosphorylation modifications 4D-DIA unlabeled |
|
| Protein phosphorylation modification PRM targeted quantification |
|
Based on a high precision and high sensitivity mass spectrometer, iTRAQ/ IBT/ TMT quantification techniques are combined with TiO2 enrichment to identify and relatively quantify modified proteins in insects, thereby investigating the relationship between dynamic changes in the modified proteome and collective phenotypic alterations in insects.
The DIA phosphoproteome is based on the unbiased mode of DIA panoramic scanning, which allows the DIA phosphoproteome to break through the limitations of traditional DDA and effectively. 4D technology enables the DIA phosphoproteome to achieve ultra-high depth identification and ultra-accurate quantification of phosphorylated proteins.
Targeted proteomic screening of target protein peptides for quantification by tandem mass spectrometry. We first screen the peptides of the target protein by primary mass spectrometry, and at secondary mass spectrometry, the PRM picks up all the fragmented daughter ion signals of the target peptides for quantitative verification of the phosphorylated modified protein of interest, primarily by LC-MS/ MS.
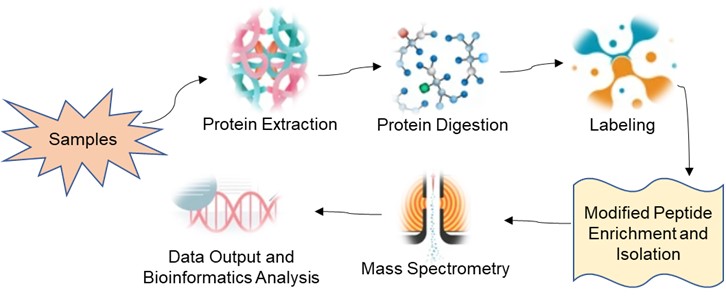 Fig 1. Service flow for protein phosphorylation modification labelling quantification.
Fig 1. Service flow for protein phosphorylation modification labelling quantification.
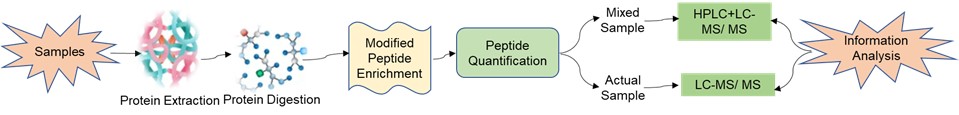 Fig 2. Service flow for quantification of protein phosphorylation modifications 4D-DIA unlabelled.
Fig 2. Service flow for quantification of protein phosphorylation modifications 4D-DIA unlabelled.
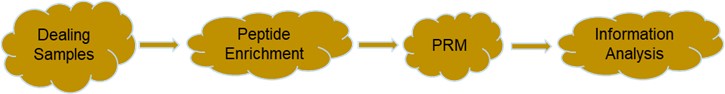 Fig 3. Service flow for protein phosphorylation modification PRM targeted quantification.
Fig 3. Service flow for protein phosphorylation modification PRM targeted quantification.
| Sample | Requirements | |
| labelling quantification PRM targeted quantification |
4D-DIA unlabelled quantification | |
| Fresh animal tissue dry weight | >20 mg | >40 mg |
| Protein solution | >1 mg (0.5 µg/ µL) | >2 mg |
| Number of freshly cultured cells | >2×107 | >4×107 |
Lifeasible provides phosphorylation modification analysis services for insect proteins to quantify the phosphorylation of insect proteins through three means. If you are interested in our services or if you have any questions, please click online inquiry for more detailed information.