The domesticated silkworm, bombyx mori, has a mid-range genome size of 432 Mb, is the model insect for the order Lepidoptera, has economically important values (e.g., silk and bioreactors production), and has been domesticated for more than 5,000 years. Because of human selection, silkworms have evolved complete dependence on humans for survival, and more than 1,000 inbred domesticated strains are kept worldwide. Archaeological and genetic evidences indicate that the domesticated silkworm originated from the Chinese wild silkworm, bombyx mandarina, that is found throughout Asia, where modern sericulture and silkworm domestication were initiated.
The silkworm is a very important domesticated economic animal and the study of its genetic variation and evolution is of great importance. The sequencing of the genome of the bombyx mori is of great importance for improving the quantity and quality of silk, disease treatment and stress resistance of the bombyx mori, which can greatly improve the economic efficiency of the bombyx mori. Lifeasible provides bombyx mori genome sequencing and analysis to accelerate the genomic and biological study of bombyx mori.
Lifeasible sequenced all bases of the bombyx mori genome to determine the base sequence of its DNA, allowing for the detection of a wide range of comprehensive mutational information at the genomic level, including single nucleotide variants (SNVs), insertion deletions (InDels), copy number variants (CNVs) and structural variants (SVs) associated with bombyx mori disease, resulting in a complete profile of bombyx mori.
| Analysis content | Detail |
| SNP detection |
|
| InDel detection |
|
| SV detection |
|
| CNV detection |
|
We sequenced the genome and later created single-end and paired (PE) libraries. Raw short reads were mapped to reference genomes using the SOAP program. All reads from the complete genome were pooled and several single nucleotide polymorphisms (SNPs) were identified using SoapSNP. The accuracy of SNP calls was assessed using Sequenom. In addition to SNPs, we can also identify insertion-deletions (indels) and validate them with PCR. Structural variants (SVs) are identified using the pairwise relationship method.
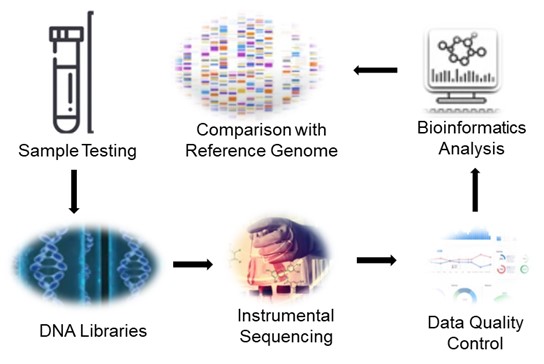 Fig 1. Service flow for bombyx mori genome sequencing.
Fig 1. Service flow for bombyx mori genome sequencing.
| Sample | Requirements |
| Peripheral blood | >3 mL, transported at 4°C. |
| Total DNA | > 3 μg, transported at 4°C. |
The five variants SNP/ InDel/ SV/ STR/ Methylation were tested simultaneously.
Lifeasible provides bombyx mori genome sequencing and analysis to accelerate the genomic and biological study of bombyx mori. If you are interested in our services or if you have any questions, please click online inquiry for more detailed information.