Drosophila melanogaster is favored by biologists for its small size, short life cycle, and ease of manipulation. It has been widely used in research in various fields, including genetics, evolution, and developmental biology. Lifeasible is committed to building three major transgenic technology platforms: transposon-mediated random insertion transgenes, integrase PhiC31-mediated targeted insertion transgenes, and CRISPR/Cas9 gene editing technology. We can efficiently and reliably design and supply transgenic Drosophila melanogaster lines for researchers in various fields, accelerating the output and translation of research results. We can also produce relevant Drosophila melanogaster disease models in high throughput to accelerate drug screening and disease research.

Indel
We generate Indel (small insertion and deletion) of genomic sequences by CRISPR non-homologous repair, which realizes code-shifting mutations in the coding regions of genes that disrupt gene function.
Knock out (KO)
We utilize CRISPR gene editing technology to cut two targets at a certain distance, and then after NHEJ repair, we realize the KO of the sequence between the two targets, which results in the loss of function of the sequence.
KI (tag)
We achieve the purpose of precise insertion of exogenous sequences through HR-mediated homologous recombination repair while cutting the target by the joint action of CRISPR and donor. It is mostly applied to inserting a tagged gene at the N- or C- terminal of the endogenous gene to analyze and obtain the gene's expression pattern.
We mainly provide two technical solutions, differentiated according to the presence or absence of a screening marker.
| Unmarked | Marked | |
| Mode of obtaining positivity | Molecular identification | Phenotypic screening (eye rfp) |
| Efficiency and success rate | High | High |
| Applicability | Wide range | Residual Loxp can't do N-terminal insertion |
| Cycle time | Short | Long (mark for removal) |
| Price | Relatively high (high cost of molecular characterization) | Relatively low (easy positive screening) |
KI (point mutation)
Under the joint action of CRISPR and donor (point mutation sequence PM), we realize the cleavage of the target while precisely replacing the original sequence of the genome by HR repair, resulting in point mutation of the coding amino acids of the gene. We optimized the design of the gRNA and donor plasmid sequences to increase the efficiency to a level comparable to that of code-shift mutagenesis.
CKOAKI (CRISPR-KO-attP-KI)
The CRISPR gene editing technology is utilized to KO the target sequence while introducing the attP site and subsequently inserting a new modified backfill sequence or exogenous sequence by the attP-PhiC31 method.
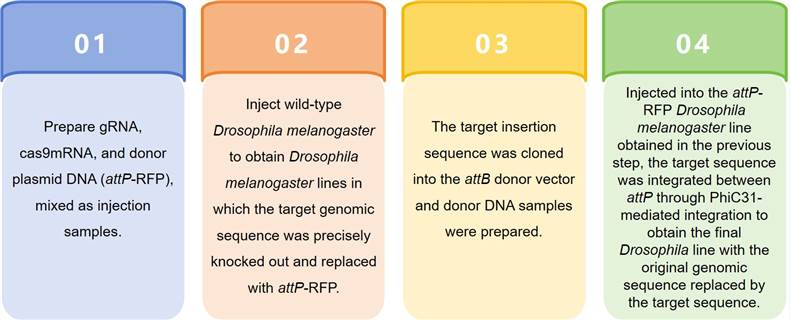
Through the CRISPR/Cas9 technological process of sample preparation, embryo microinjection, positive screening, and molecular characterization, Lifeasible can not only mutate genes in Drosophila melanogaster but also introduce exogenous fragments into the genome or knock out specific fragments with the help of donors with left and right homologous arms, realizing the precise editing of genes. Please feel free to contact us to submit your requirements.