In today's rapidly growing population, mushrooms are cultivated and consumed for their nutritional and medicinal value. The economic value of mushrooms is high, but genetic diversity is low. New strains can be generated by phenotypic selection and crosses of limited parental strains, but there is still a high degree of similarity between varieties. With current advances in biotechnology, molecular biology, and genomics, large amounts of genetic data can be generated using a variety of molecular markers that may be highly correlated with various morphological and physiological differences within the same group of mushroom species. The physiological variability, even within the same ecological niche, has increased the importance of genetic markers for studying different aspects of fungal ecology. Such molecular markers, which are stable, highly polymorphic, and provide valuable diversity information, are superior to phenotype-based markers that are influenced by the environment.
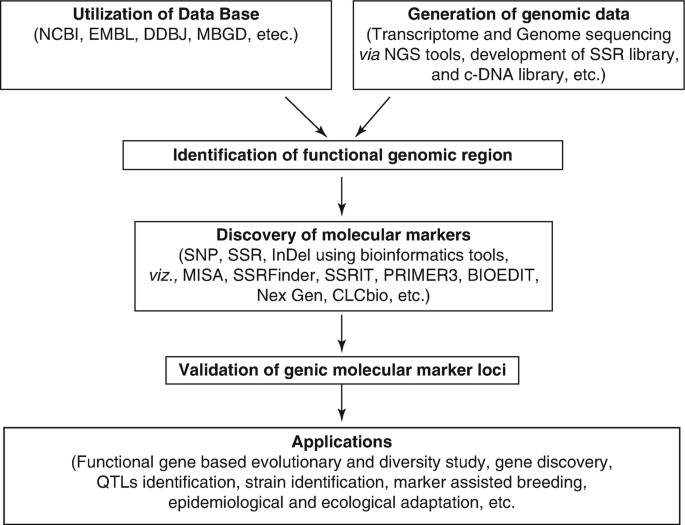 Fig. 1. Schematic presentation of the development of genic molecular markers. (Chattopadhyay A, et al., 2017)
Fig. 1. Schematic presentation of the development of genic molecular markers. (Chattopadhyay A, et al., 2017)
Services
Our team of experts continues to search for suitable genetic mutations as genetic markers for specific purposes in mushrooms. Lifeasible offers a specialized mushroom genetic, molecular markers service to evaluate the genetic characteristics of mushrooms with similar phenotypes and to support the efficient selection of mushroom germplasm, collection and conservation of strains, diversity assessment, and population structure analysis.
The large library of genome sequences enables the development of genetic markers for fungi. Lifeasible develops genetic markers for specific mushroom fungal species to establish correct taxonomy. These genetic markers are located at regulatory or functional loci and are directly linked to the trait of interest. We offer the following genetic molecular marker approaches to distinguish different mushroom species and decipher their functional diversity for marker-assisted selection to improve traits, comparative genome mapping, and exploring mushroom adaptation to different environments, etc.
- Random Amplified Polymorphic DNA (RAPD)
Our RAPD technique is widely used to assess the genetic diversity of over 37 mushroom species, and it has been observed that the technique provides better identification than morphological analysis. This technique is simple and efficient, allowing the efficient generation of many markers for genome mapping without requiring a priori sequence knowledge.
- Internal Transcribed Space (ITS) and Nuclear Large Subunit (nLSU) Markers
We provide nucleotide sequence data of ribosomal ITS regions and nLSU genes to characterize different mushroom species or strains and to identify mushroom phylogenies at the species and genus levels.
- Amplified Fragment Length Polymorphic (AFLP) Markers
We offer AFLP technology for the genomic identification of a wide range of mushrooms, such as Pleurotus ostreatus, Tricholoma matsutake, Lentinula edodes, Agaricus bisporus, and Ganoderma lucidum. This technique is precise and effective, allowing the generation and analysis of more fragments in one simple reaction, providing an almost unlimited number of genetic markers. In addition, they are stable and highly reproducible.
- Structural Variation (SV) Markers
We offer SV strategies to construct high-resolution genetic maps of mushrooms. This technique is simple, time-saving, and reproducible.
- Inter Simple Sequence Repeat (ISSR) Markers
We provide ISSR markers for genetic studies of edible mushrooms, including identification of shiitake strains, isolation of different strains of Dictyostelium, etc.
We provide a variety of genetic molecular markers to support the effective evaluation and management of new and existing mushroom germplasm, thereby facilitating further research on mushroom genetic diversity and population structure. In addition, our breeders use molecular markers for trait analysis of wild mushroom species to expand the genetic base of cultivated mushrooms. If you are interested in our services, please contact us.
Reference
- Chattopadhyay A, et al. (2017) Genic molecular markers in fungi: Availability and utility for bioprospection[J]. Molecular Markers in Mycology: Diagnostics and Marker Developments. 151-176.
For research or industrial raw materials, not for personal medical use!


 Fig. 1. Schematic presentation of the development of genic molecular markers. (Chattopadhyay A, et al., 2017)
Fig. 1. Schematic presentation of the development of genic molecular markers. (Chattopadhyay A, et al., 2017) 