Based on cutting-edge proteomics technology platforms, Lifeasible provides mushroom proteomics services to isolate and identify differentially expressed proteins in different environments and validate these proteins’ role in resisting adverse environments in combination with bioinformatics analysis.
Introduction to Mushroom Proteomics in Different Environments
In addition to their broad medicinal value, mushrooms are known for their nutritional profile and are considered an excellent source of digestible plant-based proteins. In fact, the quality of mushroom proteins is comparable to that of animal-derived protein sources and almost equal that of proteins found in meat. In people who do not consume animal proteins, mushrooms are used to combat protein deficiencies and as a supplement to grains. The protein in mushrooms contains all nine essential amino acids (EAA), whereas most other plant-based proteins are usually deficient in one or more EAA. In addition, mushrooms have a high branched-chain amino acid (BCAA) content, usually found only in animal-based protein sources. Normally growing mushrooms produce proteins specifically expressed to adapt to their environment when disturbed by external conditions such as temperature, phytohormones, salinity, and drought.
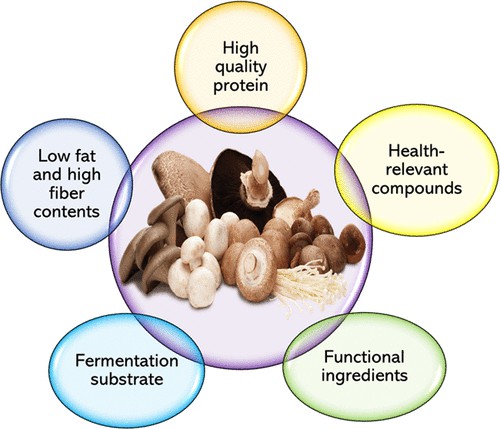 Fig. 1. Mushrooms as functional and nutritious food ingredients for multiple applications. (Kim J Y, et al., 2020)
Fig. 1. Mushrooms as functional and nutritious food ingredients for multiple applications. (Kim J Y, et al., 2020)
Services
Mushrooms are affected by the external environment and undergo changes. Our experts are very interested in the underlying mechanisms that lead to these changes. We treat mushrooms under different conditions, isolate and identify proteins extracted from treated and non-treated groups, and combine bioinformatics analysis to validate the role of differentially expressed proteins in resistance to unfavorable environments. As a leading provider of differential proteomics services for mushrooms, Lifeasible offers proteomics services for different environmental treatments.
We offer an analysis of the impact of hormones (e.g., selenosemicarbazone) added during mushroom cultivation on the proteome. Based on traditional 2-DE techniques combined with mass spectrometry, we identify differentially expressed genes in mushroom substrates.
We provide advanced iTRAQ technology to analyze the effect of cold damage on the proteome of mushroom substrates. We use SDS-PAGE to separate proteins, LC-MS to identify proteins associated with cold damage, and functional annotation and pathway enrichment to analyze the differential proteomics of mushroom substrates harvested under cold stress.
We can analyze the different manifestations of cultures at the protein level by performing proteomic analysis of mushroom mycelium in normal medium and medium supplemented with maltose and urea. We provide TOP-LC-MS/MS mass spectrometry and 2-DE techniques to isolate differentially expressed proteins from protein profiles and fully annotate their genomic information.
We use the 2-DE protein isolation technique combined with MDLDI-TOF/MS identification technique to screen and identify differentially expressed proteins in blue light-irradiated mushroom mycelium.
The differences in protein expression reflect, to some extent, changes in mushrooms in response to environmental changes. We focused on analyzing the changes of the mushroom proteome in response to different treatments, which can help to provide more accurate evaluation criteria for the genetic breeding of mushrooms based on genomics and transcriptomics and identify strains with excellent stress resistance. If you are interested in our services, please contact us.
Reference
- You S W, Hoskin R T, Komarnytsky S, et al. (2022) Mushrooms as functional and nutritious food ingredients for multiple applications[J]. ACS Food Science & Technology. 2(8): 1184-1195.
For research or industrial raw materials, not for personal medical use!


 Fig. 1. Mushrooms as functional and nutritious food ingredients for multiple applications. (Kim J Y, et al., 2020)
Fig. 1. Mushrooms as functional and nutritious food ingredients for multiple applications. (Kim J Y, et al., 2020)