As sequencing technologies continue to advance and costs continue to fall, more and more fungal genomes are being sequenced. Fungal comparative genomic analysis is based on fungal genome mapping, and sequencing technologies to comparatively analyze the structural and functional gene regions of a fungal strain genome or fungal community genome. The fungal comparative genomic analysis uses bioinformatic methods to compare fungal genomic features and then to study the evolutionary relationships between fungal species, the time of differentiation, etc. Comparative genomics has gradually become one of the essential research elements for every fungal species, especially for the first deciphered genome. Lifeasible reveals the complexity and diversity of fungi in the evolutionary process through fungal comparative genomics analysis, helping clients to elucidate the molecular basis of fungal evolution and explore the mechanism of fungal genetic origin.
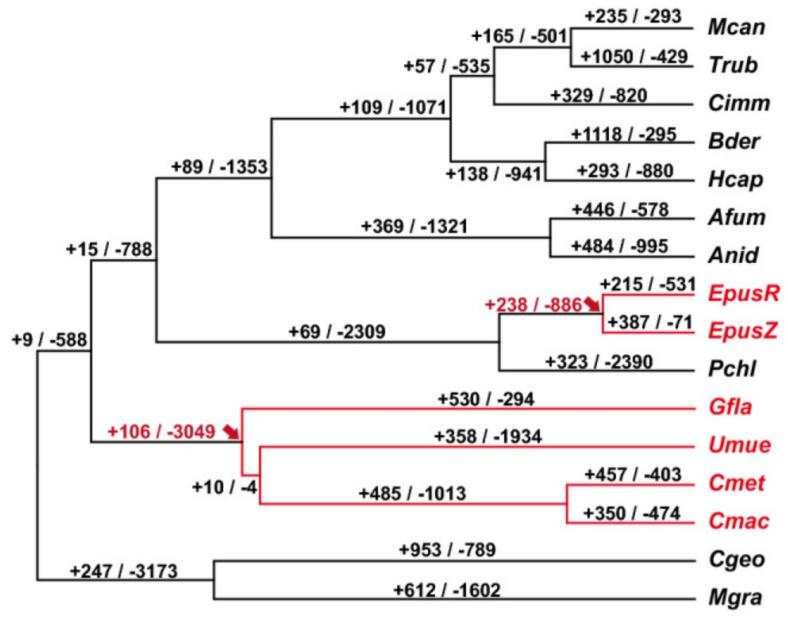 Figure 1. CAFE-based computational amplification and contraction of fungal gene families. (Song H, et al., 2022)
Figure 1. CAFE-based computational amplification and contraction of fungal gene families. (Song H, et al., 2022)
- Clustering analysis of fungal gene families.
A fungal gene family is a group of genes originating from the same ancestor and consisting of two or more copies of a single gene generated through gene duplication and interspecific divergence, which share significant structural and functional similarities and encode similar protein products. We can provide fungal gene family clustering analysis services by filtering the set, comparing the similarity of the obtained protein sequences, clustering the results, and other processes.
- Estimation analysis of the divergence time between fungal species.
Species divergence time estimation has been a challenging and topical task in studying the evolutionary history of the Tree of Life species. Species divergence time estimation in fungi is to estimate the divergence time of other taxa in the phylogenetic tree based on the divergence time between known taxa in the phylogenetic tree or based on the estimation of the base substitution rate of specific taxa by authoritative research species. We have professional bioinformatics experts who can provide estimates of divergence times for fungal species based on mcmctree.
- Fungal gene family contraction and expansion.
Determining the genetic variation behind phenotypic differences between fungal species and the evolutionary pressures leading to the changes is one of the main goals of evolutionary biology. Fungal genome analysis efforts have revealed frequent gains and losses of members of fungal interspecific gene families. Changes in the size of fungal gene families may be beneficial, detrimental, or neutral. Still, changes in the number of fungal gene families are also important reasons for species-specific formation. We can provide an analysis of fungal gene family expansion and contraction based on Computational Analysis of gene Family Evolution (CAFE).
- Fungal select pressure analysis.
Fungal selection pressure analysis refers to the pressure exerted by the outside world on fungi's evolutionary process, allowing them to adapt to their natural environment. We provide the fungal selection pressure analysis by over-analyzing the omega finger, which is the ratio of non-synonymous mutant base substitution rate to synonymous mutant base substitution rate.
- Fungal whole-genome replication events.
Polyploidy or whole-genome doubling events, which duplicate all genes within a genome, provide the original genetic material for biological evolution and are considered a gas pedal of evolution. Fungal whole genome duplication events are essential drivers of quantitative changes, formation of new gene functions, and gene rearrangements in the fungal genome. We determined the timing of fungal genome-wide ploidy through the analysis of fungal genome replication events.
Lifeasible can provide genomic analysis of fungal specimens to help clients gain insight into the mechanism of fungal gene origin and study the relationship between gene sequence and function from a gene evolutionary perspective. As your trusted partner, we can meet your fungal phylogenetic analysis needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference
- Song H, et al. (2022). "A comparative genomic analysis of lichen-forming fungi reveals new insights into fungal lifestyles." Sci Rep. 12(1): 10724.
For research or industrial raw materials, not for personal medical use!
 Figure 1. CAFE-based computational amplification and contraction of fungal gene families. (Song H, et al., 2022)
Figure 1. CAFE-based computational amplification and contraction of fungal gene families. (Song H, et al., 2022)