Many proteins are subjected to post-translational modifications (PTMs), a process where following protein biosynthesis, certain chemical moieties are attached to proteins by various enzymes. The most common and well-studied PTMs include phosphorylation, acetylation, glycosylation, ubiquitination, nitrosylation, methylation, amidation, hydroxylation, sulfation, proteolysis, etc.. PTMs can influence the protein’s physical and chemical properties, conformation, stability, and activity. Therefore, PTMs play critical roles in numerous biological processes in plants.
Lifeasible, as the world’s leading plant biotechnology company, provides high-quality protein post-translational modifications analysis. Our laboratories are equipped with advanced tandem mass spectrometry, with the capacity of obtaining high-resolution, accurate-mass (HRAM) data. Within a mass spectrometer, the protein samples are gas-phase ionized, separated according to the mass-to-charge ratio (m/z) and then quantified. Comparison of the experimentally detected intact molecular mass of purified proteins with the calculated mass obtained from the amino acid sequence of the protein will reveal any discrepancies, and the presence of PTMs can be detected by the mass difference (Δm) between unmodified and modified forms (Figure 1).
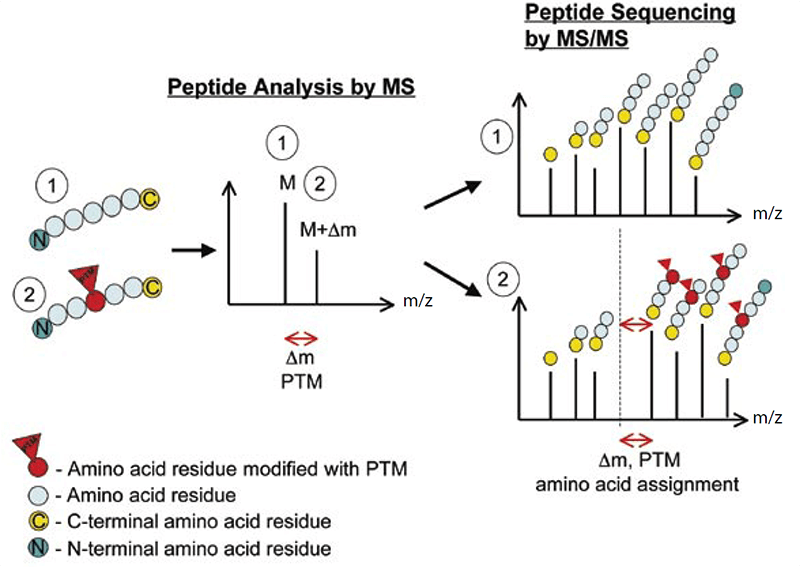 Figure 1. The principle of detecting PTMs with tandem mass spectrometry (Larsen et al., 2006)
Figure 1. The principle of detecting PTMs with tandem mass spectrometry (Larsen et al., 2006)
With cutting-edge equipment and expertise in protein characterization and mass spectrometry, we can provide you with customized protocols for PTMs analysis from both bottom-up and top-down approaches (Figure 2). The bottom-up approach requires proteolytic digestion of proteins into small peptides prior to MS analysis and is routinely used for specific protein modification identification with the partial sequence coverage and quantification with high throughput and automation. The top-down approach analyzes whole proteins directly, providing a “bird’s eye” view of all existing modifications.
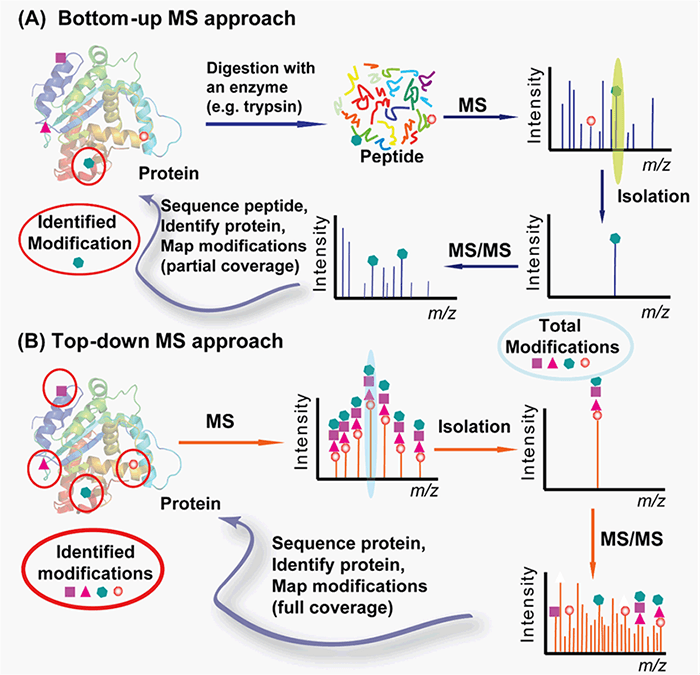 Figure 2. Top-down (A) vs. bottom-up (B) for protein PTMs characterization (Zhang and Ge, 2011)
Figure 2. Top-down (A) vs. bottom-up (B) for protein PTMs characterization (Zhang and Ge, 2011)
We offer our clients worldwide one-stop services for protein PTMs analysis (Figure 2) that cover:
Working closely with Lifeasible, you will benefit from our fast, reliable, and cost-effective services for the analysis of protein PTMs. For further information, welcome to contact us.
References