The limited morphological features and the complex evolutionary level of fungi pose significant difficulties for fungal classification. It is difficult to classify fungal species and identify their relatives by relying only on the external morphology, internal structure, and physiological and biochemical indicators of fungi. With the development and application of molecular biology, bioinformatics, genomics, and comparative genomics technologies, a revolutionary breakthrough in fungal classification have been achieved. Lifeasible can combine gene sequencing with bioinformatics technology to provide genotyping services for plant and crop fungi.
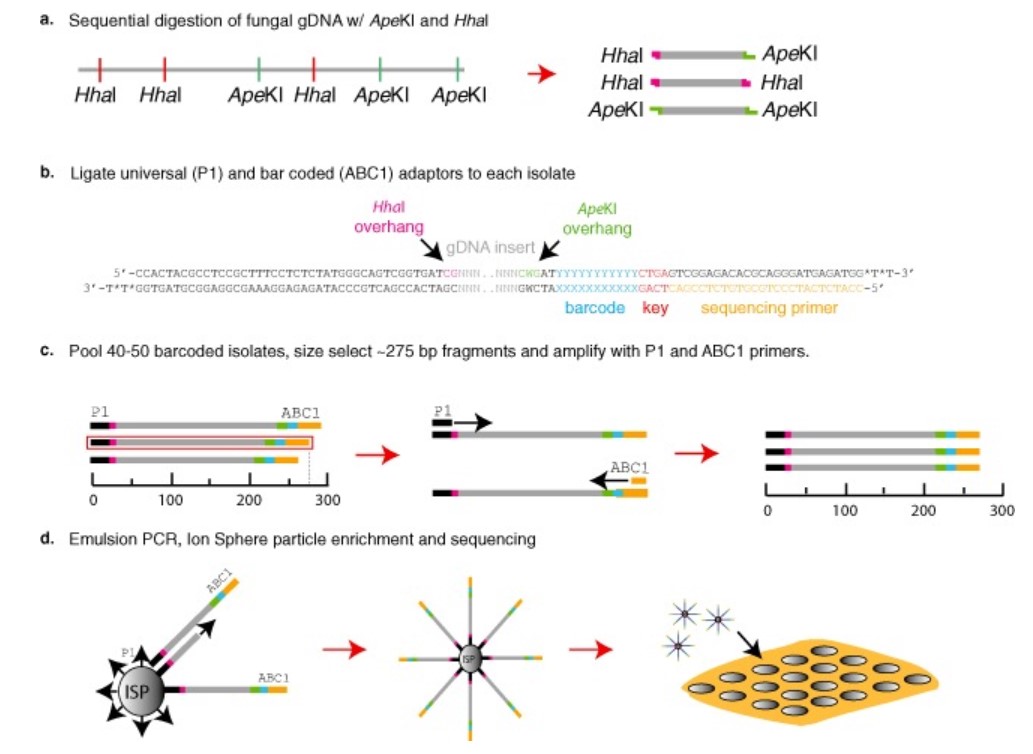 Figure 1. Genotype by sequencing (GBS) protocol for fungal genomes. (Leboldus JM, et al., 2015)
Figure 1. Genotype by sequencing (GBS) protocol for fungal genomes. (Leboldus JM, et al., 2015)
- Genotyping of fungi based on ribosomal DNA sequence analysis.
The essential difference in biological individuals is the difference in DNA sequences. The identity of two strains is determined based on whether their genomes have the same DNA sequence. There are four types of genes encoding ribosomes in the fungal genome. In general, 18S rDNA and 28S rDNA, conserved region sequence analysis is suitable for identification at the genus level, while internally transcribed spacer is ideal for identification below the species level.
- Genotyping of fungi based on microsatellite polymorphism analysis.
Microsatellite polymorphism analysis, also known as simple sequence repeat analysis, occurs in different numbers and frequencies in the genome, is widely distributed, and is highly polymorphic. We were able to genotype fungal strains obtained by isolation.
- Genotyping of fungi based on enzymatic cleavage for simplified genome sequencing.
Soil fungi are important members of the ecosystem, and toxic fungal communities invade plants through the soil and cause harm to their growth. We can extract total soil microbial DNA by amplicon sequencing technology to analyze the diversity of soil fungal community structure.
- Genotyping of fungi based on random amplified polymorphic DNA analysis.
Random amplification polymorphic DNA analysis uses a single primer composed of a random arrangement of base sequences to perform PCR to amplify all gene fragments complementary to it so that the primer and the DNA template combine to form an amplification product in the event of a possible mismatch of one or two bases, producing a series of DNA fragments of different sizes and obtaining a characteristic DNA fingerprint. We can perform fungal genotyping without primer design using RAPD techniques.
- Genotyping of fungi based on repeat sequence PCR.
Repeat sequence PCR is a DNA sequence-based genotyping tool. It uses the repeat sequence of the target gene to design two primers for PCR amplification of specific sequences. It analyzes the differences in the genome between fungal strains by comparing the results of electrophoresis of PCR products.
- Genotyping of fungi based on multi-locus sequence analysis.
Multi-locus sequence analysis reveals allelic mutations in conserved genes by sequencing for typing and identifying fungi. We analyzed the nucleotide sequences of the internal fragments of multiple housekeeping genes, assigned allele sequence numbers to all alleles of each housekeeping gene of a strain, identified the sequence type of that even strain, and analyzed the affinity between strains by comparing sequence types.
Lifeasible can use molecular biology and genomics to obtain DNA sequencing maps of fungi and provide genotyping services for fungal strains, helping customers continuously broaden the fungal research field. As your trusted partner, we can meet your fungal phylogenetic analysis needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference
- Leboldus JM, et al. (2015). "Genotype-by-sequencing of the plant-pathogenic fungi Pyrenophora teres and Sphaerulina musiva utilizing Ion Torrent sequence technology." Mol Plant Pathol. 16(6): 623-32.
For research or industrial raw materials, not for personal medical use!
 Figure 1. Genotype by sequencing (GBS) protocol for fungal genomes. (Leboldus JM, et al., 2015)
Figure 1. Genotype by sequencing (GBS) protocol for fungal genomes. (Leboldus JM, et al., 2015)