Fungal Phylogenetic Analysis
The phylogeny of fungi is the analysis of fungi based on their established relationships. Fungal phylogenetic analysis can help customers identify the evolutionary relationships among different fungal populations, understand the relationship between ancestral sequences and their descendants, and estimate the divergence times among a set of fungal populations with a common ancestor.
Fungal phylogenetic analysis is mainly based on molecular biology and builds on the molecular clock to conduct phylogenetic studies by constructing, evaluating, and interpreting evolutionary trees. Lifeasible can provide a multi-level phylogenetic analysis of fungi, providing a phylogenetic framework for exploring the rate and pattern of fungal evolution and providing strong support for clients conducting fungal phylogenetic and taxonomic studies.
 Figure 1. Diversity of major fungal lineages. (Li Y et al., 2021)
Figure 1. Diversity of major fungal lineages. (Li Y et al., 2021)
- Phylogenetic analysis of fungi based on ribosomal RNA.
Ribosomal RNA, especially the internally transcribed spacer, large subunit, and small subunit sequences, are widely used in fungal phylogenetic analysis.
The internally transcribed spacer between fungal 18S, 5.8S, and 28S rRNAs are ITS1 and ITS2, respectively. 18S, 5.8S, and 28S rRNAs are highly conserved. At the same time, ITS, which is subject to less natural selection pressure and can tolerate more variation during evolution, exhibits sequence polymorphism in fungi, while its conserved type is within species relatively consistent within species. Therefore, ITS is widely used in the phylogenetic analysis of fungi.
The ribosomal small subunit sequence species have both conserved and variable sequences, and the conserved sequences reflect the kinship between fungal species, while the highly variable sequences reflect the differences between fungal species and are suitable for taxonomic analysis above the species level.
The large ribosomal subunit has two highly variable regions, D1 and D2, which have been shown to be useful for phylogenetic analysis at the genus level.
- Phylogenetic analysis of fungi based on the mitochondrial genome.
Fungal mitochondrial genomes have substantial species differences between species. However, mitochondrial DNA sequences are relatively conserved and easier to assemble whole genome data. Therefore, mitochondrial genomes have been used for the phylogenetic analysis of fungi.
- Phylogenetic analysis of fungi based on protein-encoding genes.
Although ITS has been widely used as DNA barcodes for fungal phylogenetic analysis, a large number of studies have shown that ITS does not provide sufficient resolution for closely related plant pathogenic fungi. Therefore, we provide some fungal phylogenetic analyses based on protein-coding genes.
RNA polymerase II consists of 12 subunits, and the RPB1 gene is the most prominent functional gene responsible for encoding RNA II polymerase with a single copy and slow evolutionary rate. RPB1, in combination with other gene fragments, has been widely used in the phylogenetic analysis of fungi.
Translation elongation factor genes are often used as molecular markers for fungal identification classification and phylogenetic scheme analysis. Among them, the EF-1α sequence is suitable for phylogenetic analysis of fungi because it has higher resolution in species identification and more interspecific variation than the ITS sequence.
-
Basic steps of fungal phylogenetic analysis
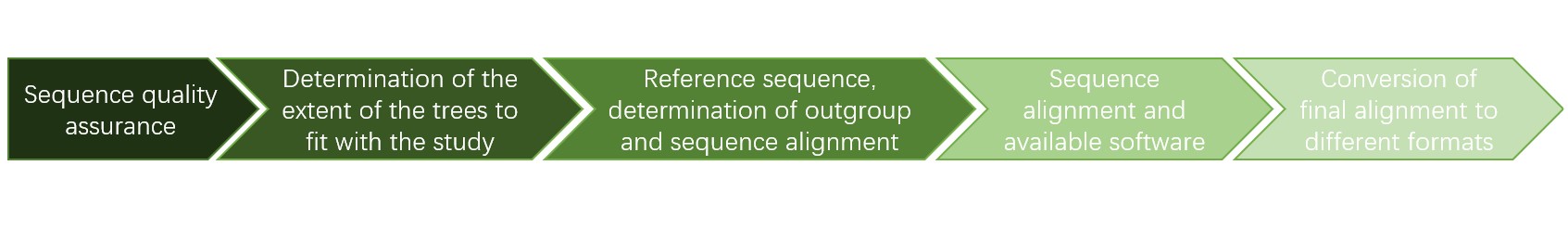 Figure 2. The workflow of fungal phylogenetic analysis.
Figure 2. The workflow of fungal phylogenetic analysis.
Lifeasible can provide fungal phylogenetic analysis based on small ribosomal subunit, large ribosomal subunit, internally transcribed spacer, and protein-coding gene sequences. As your trusted partner, we can meet your fungal phylogenetic analysis needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference
- Li Y, et al. (2021). "A genome-scale phylogeny of the kingdom Fungi." Curr Biol. 31(8): 1653-1665.e5.
For research or industrial raw materials, not for personal medical use!
 Figure 1. Diversity of major fungal lineages. (Li Y et al., 2021)
Figure 1. Diversity of major fungal lineages. (Li Y et al., 2021) Figure 2. The workflow of fungal phylogenetic analysis.
Figure 2. The workflow of fungal phylogenetic analysis.