Mammals have a strong innate immune system that works together to combat a wide range of different viruses. In addition to the interferon (IFN) response, which has been intensively studied, antiviral RNA interference (RNAi), a homology-dependent gene silencing mechanism triggered by Dicer-mediated production of small interfering RNAs (siRNAs) and microRNAs (miRNAs), is also increasingly being investigated in eukaryotes. Several studies have identified RNAi as a highly conserved mechanism of antiviral immunity in eukaryotes that plays important antiviral roles in fungi, plants, nematodes, and insects. However, previous studies have shown that Dicer activity on double-stranded RNA (dsRNA) substrates in vitro is low and that the IFN response masks or inhibits antiviral RNAi in mammals; therefore, whether RNAi contributes to the antiviral response in mammalian somatic cells remains an area of ongoing research.
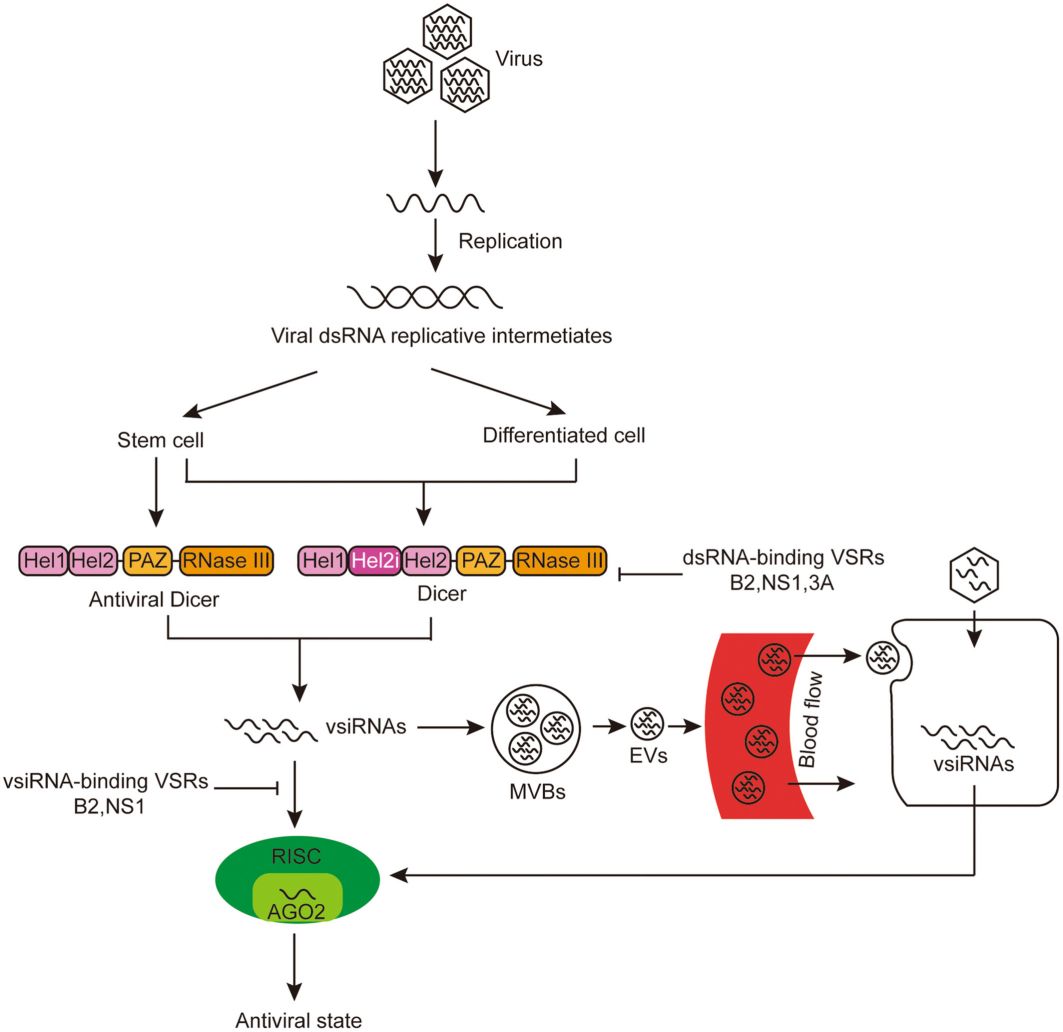 Fig. 1. Mechanism of cell-autonomous and non-cell-autonomous antiviral RNAi responses in mammals. (Wang et al., 2023)
Fig. 1. Mechanism of cell-autonomous and non-cell-autonomous antiviral RNAi responses in mammals. (Wang et al., 2023)
Lifeasible offers professional solutions to study antiviral RNA interference in mammalian cells.
Detection of dsRNAi in Mammalian Cells with Diminished IFN Response
To assess the possible presence of antiviral RNAi in mammals, it is helpful to consider studies free from virus-dependent variables. We used synthetic long dsRNAs comprising two fully complementary strands to trigger RNAi (dsRNAi). This process depends on the successive processing of long dsRNAs into a pool of siRNAs and is distinct from RNAi induced experimentally by the introduction of siRNAs (which bypasses the Dicer machinery) or of short hairpin RNAs (shRNAs, which resemble the structure of pre-miRNAs).
Analysis of Antagonism Between the IFN System and dsRNAi
We can test the antagonism between the IFN system and dsRNAi in somatic cells genetically defective in MAVS or IFNAR.
Analysis of The Efficiency of Mammalian Dicer in Processing Long dsRNA
dsRNAi in mammalian cells is further influenced by the molecular properties of its core component, Dicer. We can analyze the factors that regulate Dicer's ability to process long dsRNAs in vivo, including hDcr deletion mutants lacking almost the entire deconjugase structural domain, mouse oocytes in which dsRNAi is active express shortened Dicer isoforms. The regulation of repression by the deconjugase structural domain is likely to be achieved not through alternative transcription but through the activity of Dicer-associated proteins.
Evaluation of the Function of Antiviral RNAi in Mammals
Although knockdown of key antiviral RNAi components (e.g., Dicer and AGO2) facilitates the study of antiviral RNAi function, using Dicer-knockdown mammalian cellular models for assessing antiviral RNAi is not appropriate. We offer the following strategies to assess mammalian antiviral RNAi function.
Consistent with other aspects of immunity, antiviral RNAi is likely to be highly tunable, and it functions in conjunction with various other defense mechanisms. Lifeasible is committed to unraveling the complex network that regulates mammalian dsRNAi and to understanding its ability to act as a cell-intrinsic antiviral defense mechanism. Contact us today to learn more about our solutions.
Reference