Starch is a functionally important carbohydrate synthesized by higher plants, and starch synthase (SS) is a critical enzyme in the starch biosynthesis pathway. In recent years, scientists have been conducting bioinformatics research on plant starch synthases through informatics, computer science, and other means.
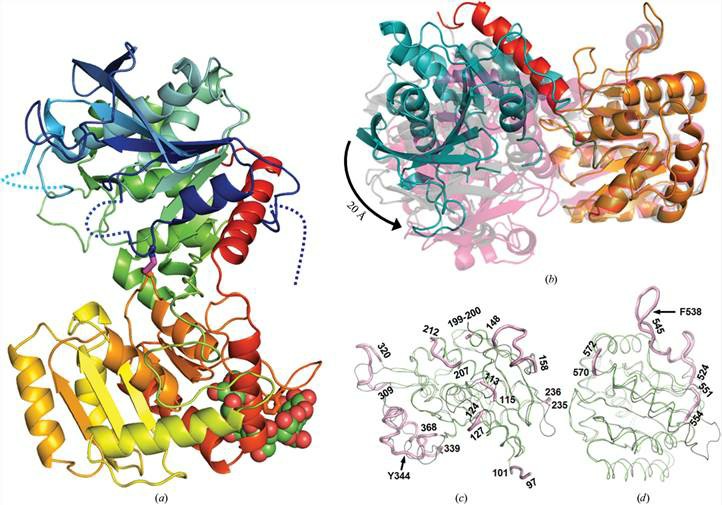 Figure 1. The structure of barley endosperm starch synthase. (Cuesta-Seijo, J. A, et al. 2013)
Figure 1. The structure of barley endosperm starch synthase. (Cuesta-Seijo, J. A, et al. 2013)
Lifeasible is a global biotechnology company with extensive experience in plant bioinformatics, covering almost all plant research services. With our outstanding bioinformatics platform, we provide our clients with bioinformatics analysis of plant starch synthases, revealing the physicochemical properties, protein structure, and phylogenetic relationships of starch synthases from multiple dimensions.
Our service mainly uses various biological analysis softwares as the main means to analyze the starch synthase of plants through query, search, and comparison to predict its physical and chemical properties, molecular structure and function, and the interaction between them.
First, we downloaded the plant's DNA sequences to be analyzed through the NCBI website and used CpG islands to analyze the methylation sites of the plant species. By determining the methylation status of the promoter CpG islands, we can understand whether a gene is expressed and provide a way to study gene expression.
We analyze the SS gene sequences of the target plants using DNAstar, ORF Finder, and ProtParam to analyze the composition and physicochemical properties of the nucleotides of the genes and their corresponding amino acid sequences.
The composition of hydrophobicity/hydrophilicity of proteins is the main driver of protein folding. We typically use ProtScale to make predictions about the amino acid sequence of plant SS.
Prediction and analysis of transmembrane structural domains are important for understanding the structure and function of proteins and their site of action in the cell. We can use TMHMM 2.0 Server to predict and analyze the transmembrane structural domains of plant SS amino acid sequences and determine their transmembrane structural domains.
We can analyze the component composition of protein secondary structures and accurately predict alpha-helices, beta-turned angles, beta-folded sheets, irregular coils, and extended chains using the PSlPRED method. Predictions can also be made for plant SS structural domains using Smart.
The Blast compares the homology of starch synthase gene sequences and amino acid sequences.
MEGA 12.0 is used to analyze the affinities between species of the target starch synthase and to construct an evolutionary tree.
| Plant Species | ||
|---|---|---|
| Solanum tuberosum | Impomoea batatas | Triticum aestivum |
| Sorghum bicolor | Cucurbita moschata | Oryza sativa |
| Eleocharis dulcis | Dioscoreaopposite Thunb. | Setaria italica |
| Musa basjoo Siebold | Solanum tuberosum L. | Sorghum bicolor (L.) Moench |
Our analysis service uses a variety of bioinformatics software to accurately identify and predict the various features of plant starch synthases, contributing to a deeper understanding of this protein.
Lifeasible specializes in the bioinformatics of plant starch synthases and provides the best possible service, from project consultation to the delivery of results professionally and scientifically. If you have any further questions, please contact us directly.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025