Nematodes are the most common, abundant, and ecologically diverse animal groups. Free-living species inhabit almost all environments and are extremely abundant in soil and aquatic sediments in freshwater and oceans. Plant nematodes attack various cash crops, mainly causing damage to the root tissue, which affects the physiological function of host plants, especially water transport. Recent transcriptome and genome projects have dramatically expanded the biological data available across plant nematodes.
Lifeasible provides a comparative analysis of plant nematode genomes, including genome sequences and mitochondrial genome sequences. Our platform is equipped with state-of-the-art facilities and highly experienced staff to assist in areas of autophagy of endoplasmic reticulum quality control. We guarantee credible results for our customers all over the world.
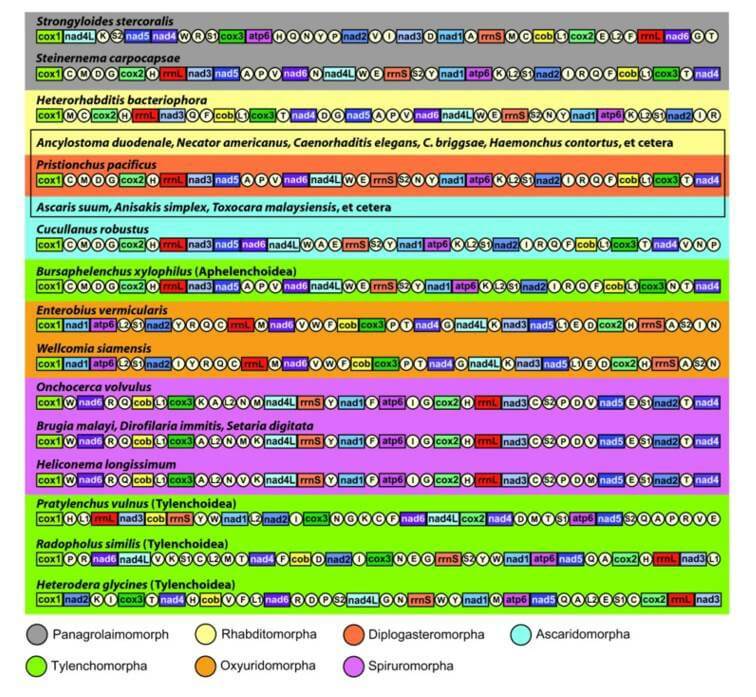 Fig.1 Linear representation of mitochondrial gene arrangement in plant nematodes. (Ma X et al., 2020)
Fig.1 Linear representation of mitochondrial gene arrangement in plant nematodes. (Ma X et al., 2020)
Lifeasible provides fast turnaround and high-quality services at competitive prices for customers worldwide. Our advanced technical platforms can help our clients solve the problems they may encounter in research based on years of experience. If you are interested in our services or have any questions, please feel free to contact us or make an online inquiry.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025