The relative abundance and broad biochemical and genetic characterization of mammalian in vitro cellular systems contrasts with the information available for other vertebrate organisms, particularly fish (the most diverse and oldest vertebrate class). Fish share many essential characteristics with mammals (e.g., similar organ systems, developmental organization, and physiological/biochemical mechanisms) and exhibit a variety of technological advantages, including adaptation to a wider range of temperatures, greater tolerance to hypoxia, and greater ease of maintaining cell cultures for extended periods. Increasingly, fish cell lines are being established from different aquaculture species and used for in vitro studies related to aquaculture and other interdisciplinary fields. Fish has become a promising vertebrate model for most biological studies and a suitable alternative to mammalian systems.
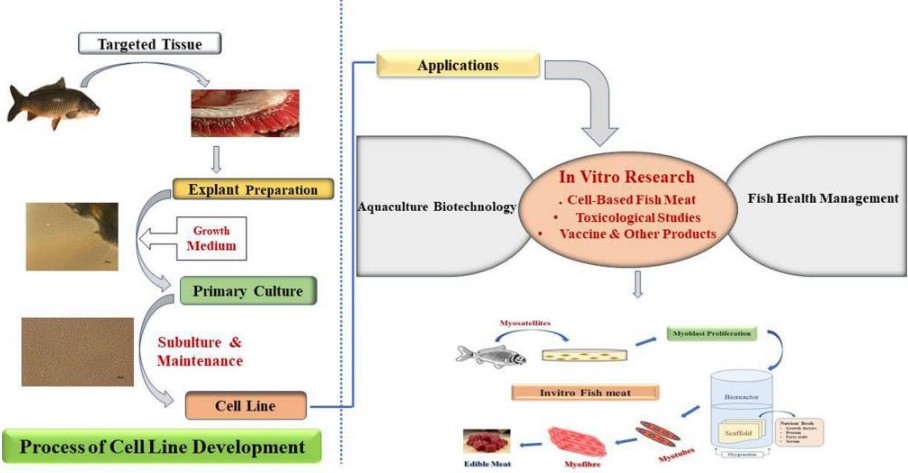 Fig. 1. Implications of fish cell line in aquaculture. (Goswami et al., 2022)
Fig. 1. Implications of fish cell line in aquaculture. (Goswami et al., 2022)
Our experts are excited in establishing and characterizing new fish cell lines representing different fish species and tissue types. Lifeasible offers customized solutions for the culture and establishment of fish cell lines. In addition, we fully characterize and validate these cell lines to ensure reproducibility without compromising scientific research.
We use different fish tissue samples (including gills, caudal fins, eyes, liver, kidneys, intestines, brain, vertebrae, and oro-nasal) to develop and establish fish cell lines. Both primary (cells isolated directly from host tissue) and established cell cultures (immortalized) are available for research. Below is a list of our available fish cell lines, including fish species and source tissues. We also offer cell cultures from marine fish.
List of cell lines developed from warm-water and cold-water fish species:
| Fish Type & Family | Species of Origin | Cell Line |
| Cyprinidae | Danio rerio (zebra fish) | ZF4 |
| Danio rerio (zebra fish) | ZEM2S | |
| Danio rerio (zebra fish) | AB.9 | |
| Danio rerio (zebra fish) | SJD.1 | |
| Pimephales promelas (fathead minnow) | FHM | |
| Pimephales promelas (fathead minnow) | EPC | |
| Cyprinus rubrofuscus (Koi carp) | EPC | |
| Cyprinus carpio (Common carp) | CCB | |
| Cyprinus carpio (Common carp) | CLC | |
| Carassius auratus (Goldfish) | CAR | |
| Poeciliidae | Poeciliopsis lucida (live bearer) | PLHC-1 |
| Clariidae | Clarias batrachus (walking catfish) | G1B |
| Ictaluridae | ctalurus punctatus (channel catfish) | G14D Suspension |
| Ictalurus punctatus (channel catfish) | 3B11 | |
| Ictalurus punctatus (channel catfish) | 28S.3 | |
| Ictalurus punctatus (channel catfish) | 42TA | |
| Ictalurus punctatus (channel catfish) | 1G8 | |
| Ictalurus punctatus (channel catfish) | CCO | |
| Ictalurus nebulosus (brown bullhead) | BB | |
| Ictalurus nebulosus (brown bullhead) | Lepomis macrochirus (blue gill) | BF-2 |
| Moronidae | Morone chrysops ( hite bass) | WBE |
| Cichlidae | Oreochromis mossambicus (Tilapia) | OmB |
| Channidae | Channa striatus (Striped snakehead) | E11 |
| Channa striatus (Striped snakehead) | SSN-1 | |
| Tetraodontidae | Fugu niphobles (Grass puffer fish) | Fugu fry |
| Salmonidae | Oncorhynchus mykiss (Rainbow trout) | RTgill-W1 |
| Oncorhynchus mykiss (Rainbow trout) | RTH-149 | |
| Oncorhynchus mykiss (Rainbow trout) | RTG-2 | |
| Oncorhynchus mykiss (Rainbow trout) | SOB-15 | |
| Oncorhynchus mykiss (Rainbow trout) | RTG-P1 | |
| Salmo salar (Atlantic salmon) | ASK | |
| Oncorhynchus kisutch (Coho salmon) | CSE-119 | |
| Oncorhynchus tshawytscha (Chinook salmon) | CHH-1 | |
| Oncorynchus nerka (Sockeye salmon) | SSE-5 | |
| Oncorhynchus tshawytscha (Chinook salmon) | CHSE-214 | |
| Oncorhynchus mykiss (Rainbow trout) | STE-137 | |
| Oncorhynchus mykiss (Rainbow trout) | TPS | |
| Oncorhynchus keta (Chum salmon) | CHH | |
| Esocidae | Esox lucius (Northern pike) | PG |
List of cell lines developed from marine fish:
| Family | Species of Origin | Cell Line |
| Gadidae | Melanogrammus aeglefinius (Haddock) | HEW |
| Sparidae | Pagrus major (Red sea bream) | SBES1 |
| Sparus aurata (Gilthead seabream) | SAF | |
| Sparus aurata (Gilthead seabream) | SaBE-1c | |
| Paralichthyidae | Paralichthys olivaceus (Olive flounder) | OFEC-17FEN |
| Scophthalmidae | Scophthalmus maximus (Turbot) | TEC |
| Lateolabracidae | Lateolabrax japonicus (Japanese sea bass) | LJES1 |
| Pomacentridae | Amphiprion ocellaris (Ocellaris clownfish) | OCF |
| Gadidae | Gadus morhua (Atlantic cod) | GML-5 |
Research and development using cell lines requires a detailed knowledge of the purity and originality of the cell line. We offer reliable solutions for fish cell line characterization based on standard protocols for characterization and identification. Some of the standard methods include cytochrome C oxidase subunit 1 (CO1) barcoding, short tandem repeat (STR) analysis, karyotyping, single nucleotide polymorphism (SNP) analysis, use of species-specific primers, whole genome sequencing (WGS), CLASTR, DNA-based authentication, etc.
Lifeasible explores fish cell lines to produce a variety of new products. We use standard protocols to maintain consistent quality and validate cell lines throughout their in vitro life cycle. Contact us today to learn more about our solutions.
Reference