Fluorescence in situ hybridization (FISH) is a valuable cytogenetic technique for the detection and localization of specific DNA or RNA sequences on chromosomes (1). In general, this technique can be used for defining the spatial-temporal expression patterns of target genes. Utilizing fluorescent DNA probes with sequences complementary to targeted chromosomal locations, the gene expression patterns can be easily detected by fluorescent microscope, and further subjected to quantitative analyses (Figure 1). Moreover, by labeling different sequence fragments with distinct fluorophores, the improved FISH assay also allows simultaneous observation of multiple genes (2). Lifeasible provides high quality service for both single- and multi-color FISH assays with improved efficiency and competitive price. Besides, new developed variants of the classical FISH assay have also been designed to largely broaden the application scope of this technique.
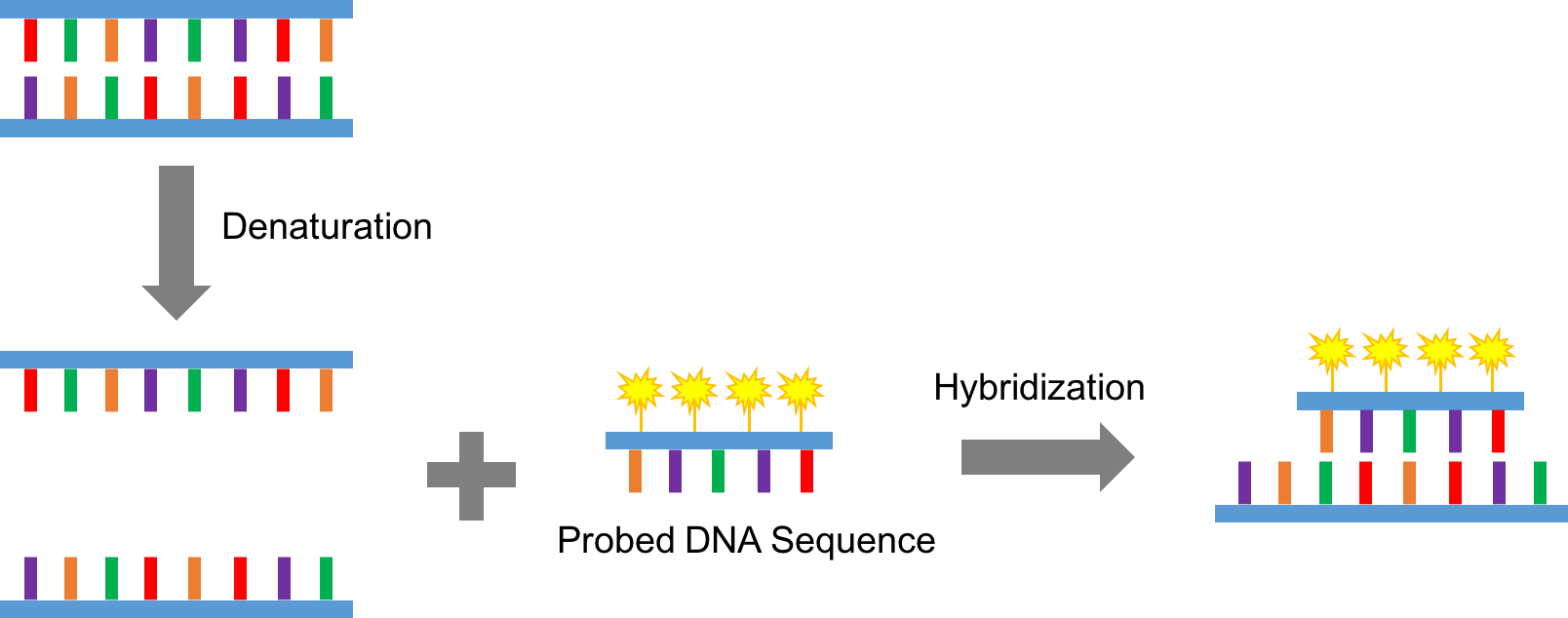 Figure 1. A schematic cartoon of the principle of FISH assay
Figure 1. A schematic cartoon of the principle of FISH assay
Compared to other in situ hybridization assays, the FISH assay has the following advantages:
In the plant system, the FISH assay has been employed in multiple crop species, including wheat, rye, cucumber and melon (3, 4). With solid biochemical and cytological backgrounds, and years of experience in plant cell line/tissue sample generation, scientists and experts here at Lifeasible provide high quality service that covers every step of the FISH assay in the plant system:
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025