With the continuous development of molecular biology, a new generation of molecular marker types have emerged, such as Sequence Tagged Site (STS), Sequence Charactered Amplified Region (SCAR), Candidate Gene (CG), etc. Sequence Tagged Site (STS), Sequence Charactered Amplified Region (SCAR), and Candidate Gene (CG) based on sequencing. The sequence tagged site (STS) is a short DNA sequence (200-500 bp) that occurs only once on a chromosome and whose position and base order are known. The greatest advantage of using STS is that it produces very reliable information.
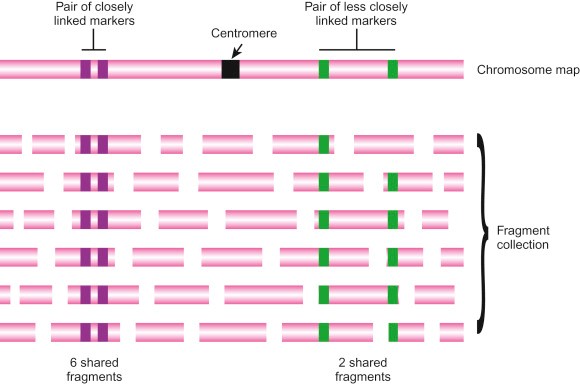 Figure 1. Mapping of Sequence Tagged Sites. (David PC, et al. 2013)
Figure 1. Mapping of Sequence Tagged Sites. (David PC, et al. 2013)
Unlike PCR with arbitrary primers, STS are primers that are based on some degree of sequence knowledge. These unique, sequence-specific primers detect variation in allelic, genomic DNA. STS have a particular advantage over RAPD in that they are codominant, that is, they can distinguish between homozygotes and heterozygotes. They also tend to be more reproducible, because they use longer primer sequences. However, they have the disadvantage of requiring some pre-existing knowledge of the DNA sequence of the region, even if only for a small amount. The investment in effort and cost needed to develop the specific primer pairs for each locus is their primary drawback. As with RAPD, using PCR produces a quick generation of data and requires little DNA. All STS methods use the same basic protocols as RAPD (DNA extraction and PCR) and require the same equipment.
Any unique DNA sequence can be used as an STS. for a DNA sequence to be an STS, its sequence must be known so that the presence or absence of the STS in different DNA fragments can be detected by PCR. the STS must be uniquely localized on the chromosome to be studied or, when the DNA fragment population covers the entire genome, the STS must have a unique locus in the entire genome. If the STS sequence has multiple localization sites, the mapping data will be ambiguous. Therefore, it is necessary to ensure that the STS does not contain sequences of repetitive DNA.
Lifeasible has many years of experience in molecular markers, we provide STS technology solutions for global customers, we have a professional team to provide you with complete plant genomic DNA extraction and purification, STS-PCR primer design screening, sequence feature amplification region, and other project processes, with experienced experts to participate in the whole project, to supervise the project completion progress and quality, aiming to provide you with the best quality STS molecular marker services.
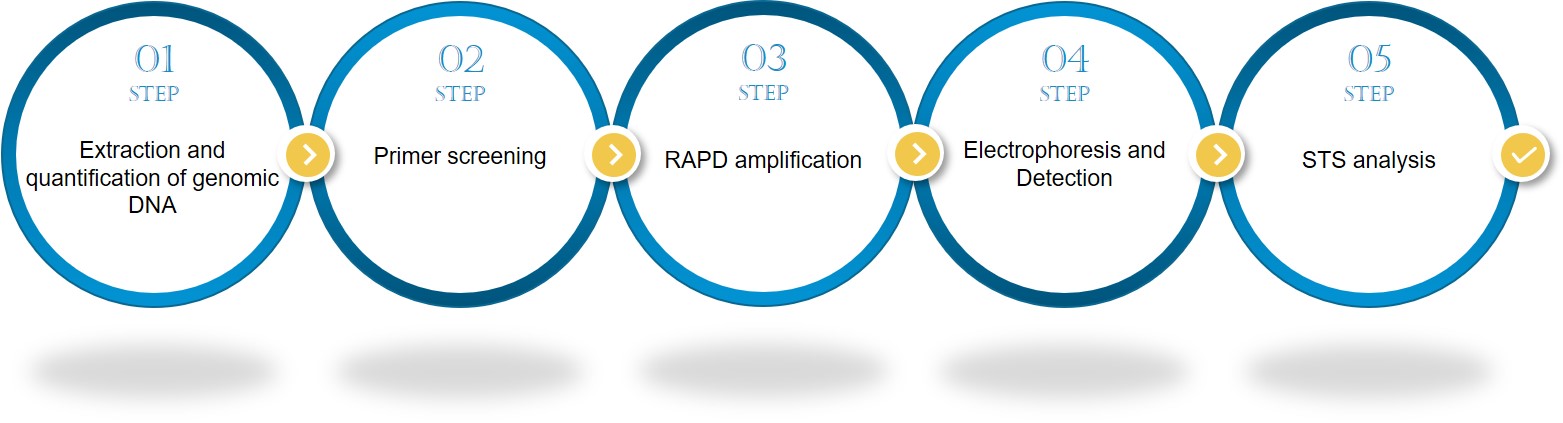

Lifeasible offers complete, professional STS technology service, as well as customized experimental protocols based on your project requirements and plant sample characteristics. For more information, please contact Lifeasible.
Reference:
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025