Forests are an essential part of the Earth's ecosystem, with about 100,000 known tree species and trees covering about 31% of the Earth's area. Most current tree species are naturally bred heterozygotes and contain a large amount of genetic information. The diversity of forest genetic information can not only ensure the variety of species, but also the reasonable study of forest genetic diversity can effectively understand the origin of tree species, estimate the distribution of genetic resources, develop and utilize high-quality tree resources, and conduct cross-breeding among high-quality tree species.
Lifeasible is deeply involved in plant molecular technology and has built a state-of-the-art plant genetic information analysis laboratory, which can perform genetic diversity analysis for a wide range of forest trees. We guarantee to provide accurate and fast forest genetic diversity analysis services to our customers worldwide.

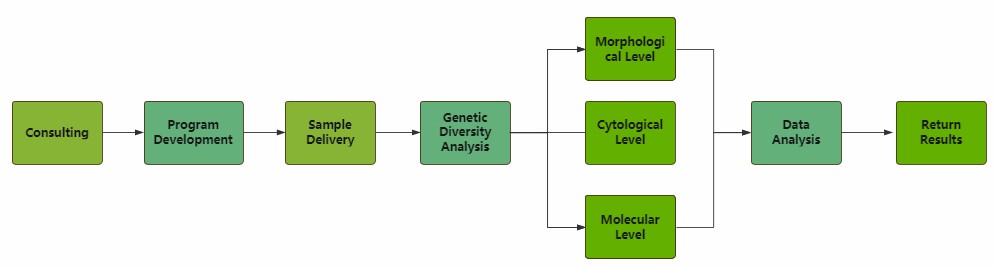
With the advancement of molecular biotechnology, we can study the genetic diversity of forest trees at more levels and help our clients to have a more comprehensive understanding of the genetic diversity of forest trees.
Our analytical services focus on three main areas.
We use the plant's external characteristics and morphological anatomy to perform markers and analysis to analyze the genetic relationships between phenotypes and genotypes.
The analysis at the cytological level is based on the karyotypic and banding characteristics of cellular chromosomes. We examine the number, size, follower, and mitotic sites of chromosomes at different times to analyze the rich genetic diversity of tree species.
The analysis at the molecular level focuses on two types of large molecules, proteins (enzymes) or DNA of forest trees. Horizontal section starch gel electrophoresis (SGE) and polyacrylamide gel electrophoresis (PAGE) can be used to analyze information such as band types of forest tree proteins. Forest DNA is then analyzed using different molecular marker methods, including random amplified polymorphic DNA, amplified fragment length polymorphism, single-stranded conformational polymorphism, small satellite simple sequence repeats length polymorphism, and simple repeat sequence interval polymorphism. Research at the molecular level has greatly facilitated the study of genetic diversity in forest trees and can well help us analyze the genetic diversity of forest trees.
Our forest genetic diversity analysis service covers a wide range of common tree species, and the following shows the information on the main forest tree species covered by our company.
Betula pendula
Pinus strobus
Pinus koraiensis
Eucalyptus grandis
Populus tremula
Populus pruinosa Schrenk
Populus deltoides Marshall
Pinus lambertiana
Ginkgo biloba
Gnetum montanum
Carya ovata
Carya illinoinensis
Acer saccharum
Quercus bicolor
Larix sibirica
Taxus baccata
Abies alba
Quercus alba
Pinus ponderosa
Pinus massoniana
Sequoiadendron giganteum
Lifeasible has built a complete plant genetic diversity testing laboratory equipped with advanced molecular sequencers and morphological testing instruments, which can provide accurate data for forest genetic diversity analysis services. Our technical team will always be in contact with you to ensure that we can solve any problems that may arise during the project. If you are interested in our projects, please contact us directly.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Mechanism of Action of Plant Resistosome NRC4
May 20, 2025
Mechanisms Regulating Plant Chloroplast Biogenesis
April 15, 2025