According to statistics, there are 1.6 million species of fungi worldwide, and about 70,000 species have been described. The fungal species are large and diverse and are widely distributed in nature. Fungal gene sequencing is the basis for fungal genomics analysis and the application of multi-omics technologies in the field of fungal research. As a leading provider of agricultural research services, Lifeasible provides professional fungal genome sequencing services and offers different solutions based on customer needs, such as second-generation sequencing. Third-generation sequencing platforms, to help customers gain a comprehensive understanding of fungal genomic information.
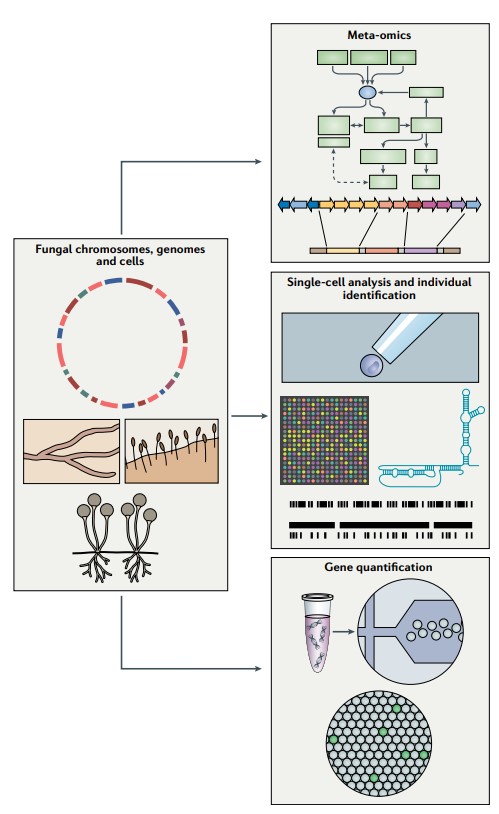 Figure 1. Emerging sequencing techniques. (Nilsson RH, et al., 2019)
Figure 1. Emerging sequencing techniques. (Nilsson RH, et al., 2019)
Fungal whole genome de novo sequencing is to obtain the whole genome sequence map of a fungus with unknown genome sequence or no genomic information of closely related species by constructing a genomic DNA library with different inserts and sequencing the library, and then using bioinformatics methods for virtue, assembly, and annotation.
Fungal genome resequencing is the high-throughput sequencing of an individual or a population of fungi with a reference genome. Based on high-throughput second- or third-generation sequencing technology to obtain the whole genome sequence of the fungus, the sequencing data are compared to the reference genome to obtain a series of variation information of the fungal individual or population relative to the reference genome. Fungal genome resequencing can help customers discover single nucleotide polymorphic, copy number variants, insertions, deletions, and other variant types and amplify individual reference genomic information into genetic traits of biological populations.
The specific services that Lifeasible can provide are as follows, including but not limited to:
| De novo sequencing of fungal genomes | Fungal genome resequencing |
| Sequencing data quality control analysis | Sequencing data quality control analysis |
| Fungal genome assembly | Consistent sequence assembly of fungal genomes |
| Gene structure prediction and function annotation | Single nucleotide polymorphism site detection |
| Gene family analysis | Analysis of gene copy number changes |
| Repeat sequence annotation | Structural variant site detection |
| Gene ontology annotation | |
| ncRNA annotation |
Lifeasible is capable of de novo sequencing and resequencing the fungi's whole genome by various sequencing technologies. The fungal whole genome second-generation sequencing technologies we can offer include pyrophosphate sequencing and Illumina sequencing. Pyrophosphate sequencing associates the polymerization of each dNTP on a primer with the release of a single fluorescent signal. Pyrophosphate sequencing is suitable for sequencing analysis of known short sequences, does not require gel electrophoresis or any special form of labeling and staining of DNA samples, and is high-throughput, low-cost, and rapid. Illumina sequencing, with its core principle of sequencing while synthesizing, is the default choice for mycological studies.
The fungal whole genome triple sequencing technology we can offer is nanopore sequencing technology, which is centered on the use of nanopores, allowing individual nucleic acid polymers to pass through the nanopore, with different bases having different charged properties, and determining the base sequence by monitoring and decoding the current signal in real-time.
Lifeasible can provide fungal whole genome de novo sequencing services and genome resequencing. As your trusted partner, we can meet your fungal whole genome sequencing needs and provide you with efficient, high-quality services. If you want to know the details, please contact us.
Reference