Plants undergo various physiological and pathological processes, and their genomes are usually always stable; however, their proteome composition changes with different environments or differences in their physiological states, forming different types of proteins. Plants respond to fungal infestation by producing new proteins, called pathogenesis-related proteins. With the increasing advancement of proteomic analysis technology and the construction of plant host-pathogenic fungal interaction system, a large number of functional genes and proteins related to pathogenesis-related and defense responses have been verified, and proteomics can be used as a complement to genomics and transcriptomics to reveal the molecular mechanisms of the plant. Lifeasible analyses of differentially expressed proteins derived from plant hosts in plant-pathogenic fungal intercropping systems with the help of established proteomic analysis methods and biomass identification techniques. We help our customers to deeply understand the complex molecular mechanisms of pathogenic fungal infestation by clarifying the specific roles of proteins in defense response mechanisms.
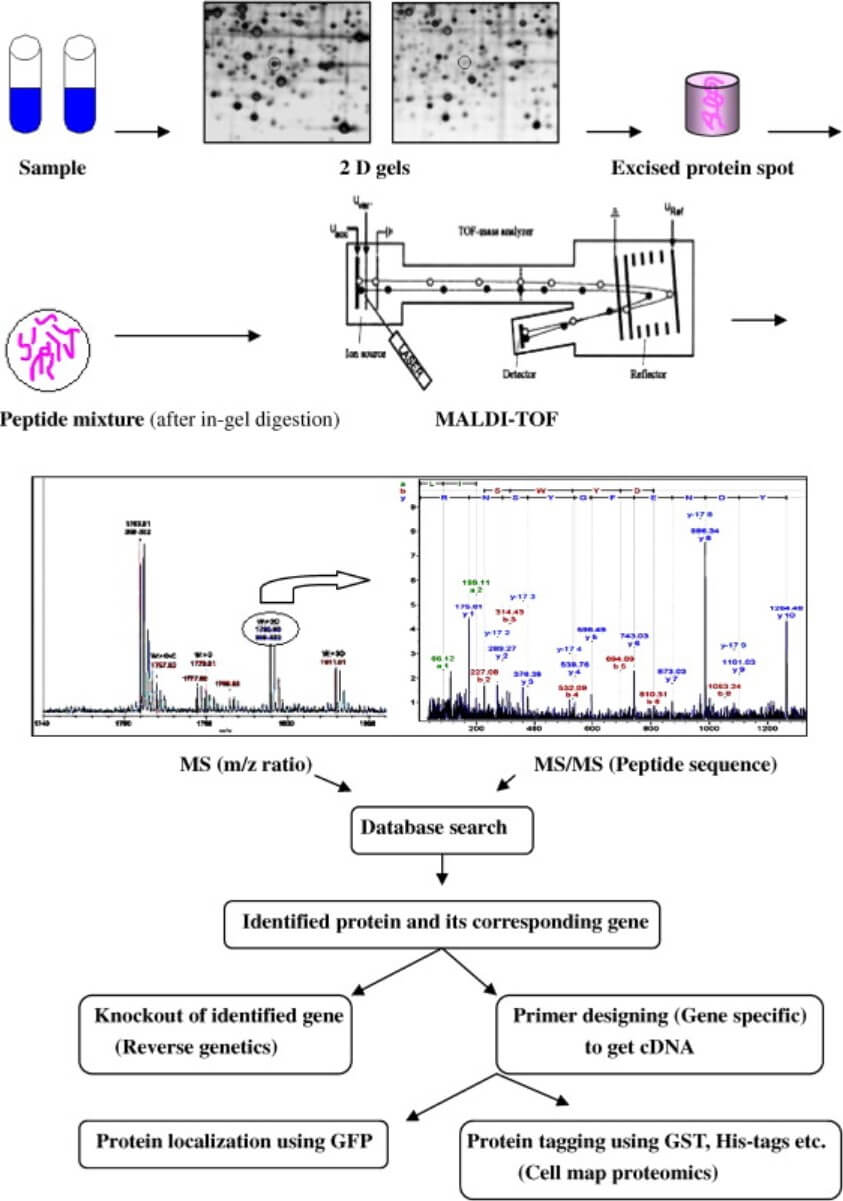 Figure 1. Principles of proteomics. (Bhadauria V, et al., 2007)
Figure 1. Principles of proteomics. (Bhadauria V, et al., 2007)
Proteomics is often used as a complementary technique to help clients gain a comprehensive understanding of the changes that occur in protein expression during plant-fungal interactions. We typically use two-dimensional gel electrophoresis to construct proteomic profiles, identify and characterize proteins, and analyze proteomic expression differences between disease-sensitive and corresponding normal tissues by differential protein expression.
Plant pathogenic fungi are capable of secreting many different types of effector molecules, including effector proteins, small RNAs, phytohormones, and their derivatives. The secretory proteome refers to all proteins secreted by tissues or cells. As a branch of proteomics, secretory proteomics is studied based on existing proteomics research techniques and methods. We can analyze the important functions of secreted proteins of different lifestyle fungi in the communication between fungi and plants using bioinformatics and molecular biology techniques, among others.
| Commonly Used Software for Secreted Protein Prediction |
| Signal Peptide Prediction Software SignalP v4.1 |
| Signal Peptide Prediction Software SigCleave |
| Signal Peptide Prediction Software RPSP |
| Subcellular localization Position Prediction Software TargetP |
| Subcellular localization Position Prediction Software WoLF PSORT |
| Transmembrane Helix Structure Prediction Software TMHMM |
| Transmembrane Helix Structure Prediction Software Phobius |
Lifeasible can help customers explore changes in protein expression levels in plant-fungal interaction systems using various proteomics research techniques. As your trusted partner, we can meet all your fungal proteomics analysis needs and provide you with efficient and high-quality services. If you want to know the details, please contact us.
Reference