The implementation of plant biotechnology makes it possible to modify crop traits in a targeted manner through genetic transformation. Insertion of specific transgenic elements containing a desired gene into the host genome is by far the most common application of this technology. Safety assessment of new genetically modified plants is critical to identify potential threats to humans, animals, and the environment. The first step in risk assessment of a genetically modified plant before it is allowed to be placed on the market is the molecular characterization of the genetically modified plant. Molecular characteristics refer to the integration of foreign DNA fragments in transgenic plants into the recipient genome, including the copy number of the inserted foreign gene, the flanking sequences of the insertion site, the integrity of the inserted sequence, whether there is insertion of the vector backbone sequence, and expression and information on genetic stability, and other aspects. Information on these characteristics is the basis for developing, safely assessing, testing, and monitoring genetically modified crops.
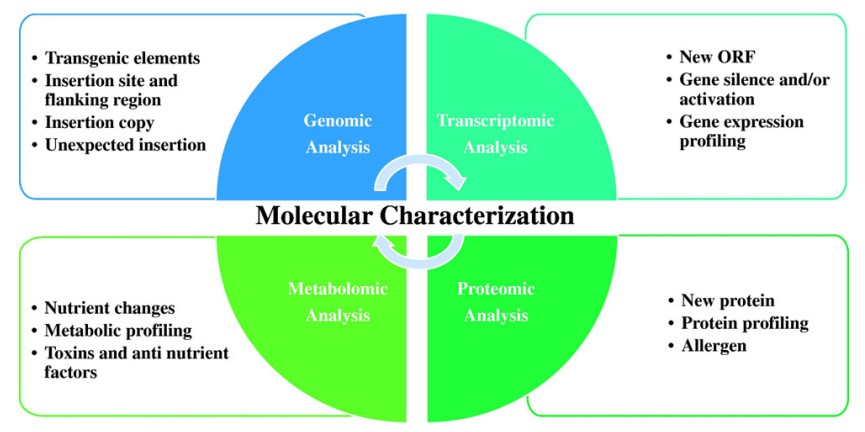 Fig. 1. Molecular characterization of genetically modified crops at genomic, transcriptomic, proteomic, and metabolomic levels. (Li et al., 2017)
Fig. 1. Molecular characterization of genetically modified crops at genomic, transcriptomic, proteomic, and metabolomic levels. (Li et al., 2017)
Molecular characterization analysis of transgenic plants is important in determining whether the target gene has been successfully transformed into the plant. Based on its rich experience in the field of genetically modified plants, Lifeasible provides customers with molecular characterization analysis services for a variety of genetically modified crops with stable quality and reasonable prices, including but not limited to:
We have matured genetically modified plant screening and detection technology. Molecular characterization analysis of transgenic plants can be performed at all levels (DNA, RNA, and protein). Our detection technologies include:
1. DNA Level
This method has the advantages of high efficiency, simple operation, and low cost. When used to analyze transgenic plants, if the gene of interest is successfully transformed into the plant, a band of a specific size will appear in the gel.
This method uses labeled probes to specifically hybridize to target DNA and has high accuracy and specificity. For molecular characterization of transgenes to determine the presence and copy number of introduced genes into the GMO genome. We have successfully applied this technology to screen genetically modified plants in various crops such as rice, wheat, corn, soybeans, rape, and peaches.
2. RNA levels
In transgenic plants, RT-PCR detects whether the foreign gene is expressed at the mRNA level through electrophoresis. RT-PCR can detect low-abundance mRNA in transgenic plants, especially when the target gene is integrated into a single copy.
Northern blotting can be used to analyze the size and level of target RNA in transgenic plants and whether there are relatively large amounts of gene expression at the RNA level. We can use this technology to study the expression and regulation of foreign genes in transgenic plants such as potatoes, rice, and tobacco.
It is a method for relatively quantitative analysis of exogenous gene copy numbers. Its main feature is to monitor the entire PCR process through fluorescence signal accumulation. It is helpful to detect the relative expression of mRNA in transgenic plants. It has the characteristics of high sensitivity, high accuracy, and a high degree of automation for screening transgenic plants.
3. Protein Levels
This method is used to qualitatively analyze target proteins in transgenic plant samples. Based on the test results, we can know whether the target protein is expressed in the transgenic plants, its concentration, and its approximate molecular weight. We have widely used this technology to detect protein expression in genetically modified plants such as tobacco, wolfberry, and poplar.
It provides highly sensitive information on the protein content in genetically modified plant samples. We have widely used this technology to screen cotton, pepper, rice, tobacco, tomato, and other transformed plants.
4. High-throughput sequencing technology
We have multiple sequencing platforms that can be used for transgene sequencing analysis, including Roche/455, Illumina HisSeq, Solid, Heliscope, and third-generation sequencing platforms PacBio SMRT, Nanopore nanopore sequencing, etc.
By sequencing the genome of genetically modified organisms and then using the vector DNA sequence to capture the exogenous inserted DNA sequence and the binding region sequence between exogenous DNA and the receptor genome from the sequencing data, the molecular characteristics of the genetically modified organism are analyzed based on the binding region sequence. This method can be used to analyze the flanking sequences of exogenous inserted sequences in transgenic organisms and can also explore the copy number of exogenous inserted sequences.
A technology that fragments genomic DNA uses probes (designed based on the known sequence of the transformation vector) to capture target DNA fragments, enriches them, and then performs sequencing. We have two technologies, T-DNA capture sequencing and Southern by sequencing (SBS) capture sequencing, to determine the integration of foreign genes in the genome of genetically modified organisms.
Lifeasible is committed to providing comprehensive molecular characterization analysis services for genetically modified plants. We will select appropriate molecular characterization analysis methods for different species based on your different needs to ensure objective and reliable results. If you have any questions, please feel free to contact us. We look forward to working with you on projects of interest.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025