Lifeasible is dedicated to developing RNA interference technology in plant protection and provides identification and validation services for small RNAs that can be used for plant protection.
RNAi-based approaches can significantly contribute to integrated pest and pathogen management and sustainable development of agriculture. The first step in using RNA interference to protect plants is to identify RNA interference-associated small RNAs (sRNAs) and determine the silencing targets of sRNAs. The development of second-generation sequencing technologies has accelerated the sequencing of sRNAs and sRNA targets, providing basic expression information and expression differential information. Then, computer-based information analysis also facilitates the identification, clustering, and functional prediction of sRNAs. Furthermore, multi-validation methods of sRNAs help to determine the exact role of sRNAs. Lifeasible makes full use of advanced sequencing technology, comprehensive bioinformatics analysis, and multiple sRNA validation methods to help our customers identify and validate sRNAs for plant protection.
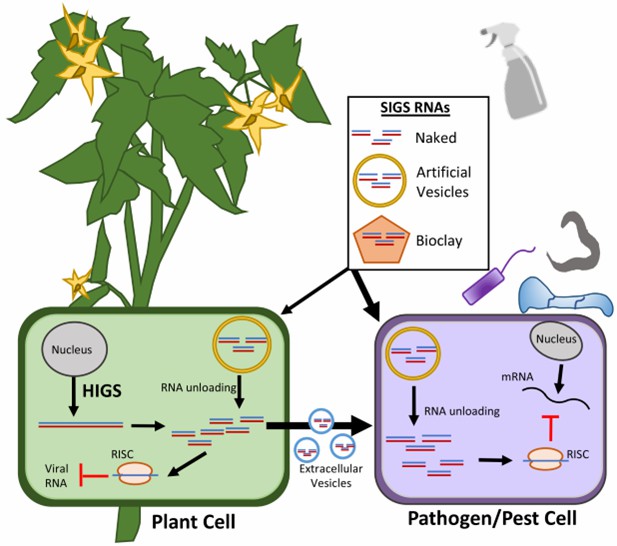 Fig. 1 RNAi-based plant protection strategies (Niu et al., 2021).
Fig. 1 RNAi-based plant protection strategies (Niu et al., 2021).
We provide targeted sRNA identification services.
Table. 1 Identification of sRNAs
| Subjects | Specific sRNA identification services |
| Plants |
|
| Pests |
|
| Pathogenic fungi |
|
| Plant viruses |
|
Computer-based sRNA prediction
We can analyze existing genetic information, transcriptomic data, and RNA libraries and predict sRNAs that can be used for plant protection. We have developed an enhanced sRNA scanning method to predict sRNAs efficiently.
Advanced sequencing technology
We use advanced third-generation sequencing and next-generation sequencing technologies to detect nucleic acid sequences for efficient mining of sequence information. We mainly use deep sequencing with Illumina technology for the sequencing of sRNAs.
Comprehensive bioinformatics analysis
Our bioinformaticians can perform a comprehensive analysis of the nucleic acid information. For example, when analyzing RNA data, they use bioinformatics to identify sRNAs present in RNA databases, to predict miRNAs that are not identified, to do hierarchical clustering analysis of differentially expressed miRNAs, to perform multilayered enrichment analysis (GO and KEGG annotation).
Multiple sRNA validation methods
For computer-predicted sRNAs, we will perform sequencing to validate them. For the validation of miRNAs, we mainly use qRT-PCR for validation. We also offer validation using chemically synthesized dsRNAs that then act on the target organism. If there is a demand for cloning validation, we can also provide it.
Lifeasible provides professional sRNA identification and validation services, please contact us to serve you.
References
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025