RNA editing is a post-transcriptional processing modification of chloroplast genes in higher plants that alters the genetic information carried on mRNA through base insertions, deletions, and substitutions, correcting mutations that occur at the DNA level and directing the synthesis of functional proteins. Chloroplast RNA editing affects chloroplast biosynthesis, and inhibition of chloroplast biosynthesis will affect plant photosynthesis, resulting in adverse consequences such as plant yellowing, dwarfing, prolonged flowering, and delayed fruit ripening. Given this, research on RNA editing in plant chloroplasts is essential.
Lifeasible provides services for identifying and analyzing plant chloroplast RNA editing loci to help our clients elucidate the mechanisms and properties of RNA editing in the plant chloroplast genome. We have always adhered to the philosophy of "scientific, realistic, efficient and rigorous", and we are committed to providing one-stop biological services to meet the needs of our clients.
RNA editing is dominated by cytosine (C) to uracil (U) substitution in plant chloroplasts and may be an important way for chloroplasts to produce functional proteins. In recent years, techniques such as in vitro analysis, chloroplast transformation, and UV cross-linking have often been used to probe the mechanism of chloroplast RNA editing. We focus on the composition and characterization of RNA editing sites to provide solutions for our clients.
The combination of bioinformatics software and experimental validation is our service strategy. We first use isolation and purification of plant organelles to obtain chloroplasts, extract total RNA from chloroplasts and construct libraries for sequencing. Bioinformatics tools such as SAMtools, BDD tools, and ChloroSeq are then used to predict RNA editing sites for plant chloroplast. Finally, the predicted loci were experimentally validated using RT-PCR combined with clone sequencing.
We have provided our clients with several successful identifications of chloroplast RNA editing sites. We can target almost all plant species with this service, including but not limited to those shown in the table below.
| Our successful project experience | ||
|---|---|---|
| Anthoceros formosae | Oryza sativa | Nicotiana tobacum |
| Adiantum capillus-veneris | Saccharum officinarum | Arabidopsis thaliana |
| Zea mays | Pisum sativum | Solanum lycopersicum |
| Phalaenopsis Aphrodite | B. pendula | Corylus chinensis |
| Juglans sigillata | Setaria italica | Betula |
| Lavandula angustifolia | Ocimum basilicum | Mentha canadensis Linnaeus |
| Leonurus japonicus Houtt | Rosmarinus officinalis | Perilla frutescens |
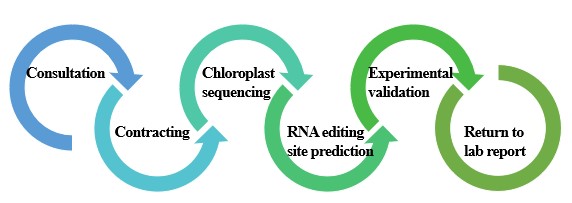
Lifeasible, as a professional plant biotechnology company, has focused on plant chloroplast-related research for more than a decade. We have built an advanced plant biotechnology platform and can provide a one-stop service from organelle isolation to chloroplast RNA editing site identification. If you are interested, please feel free to contact us for details and a quotation.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025