Label-Free Proteomics Analysis
Proteomics is a core technology to systematically study the expression, function, and regulation of all organisms' proteins. As a quantitative method without chemical or isotopic labeling, label-free proteomics directly analyzes the mass spectrometry signals of proteolytic peptides by liquid-liquid-mass spectrometry (LC-MS/MS). It achieves relative quantification based on the peak intensity or peak area. Compared with labeled techniques, label-free methods have the advantages of low cost, simple sample processing, and wide applicability, especially suitable for studies with large numbers and complex types of samples.
Lifeasible has rich experience in label-free proteomics analysis, and our advanced technology platform and professional technical team aim to provide high-quality and accurate label-free proteomics services.

The Significance of Label-Free Proteomics
- Technical simplicity. Label-free proteomics methods are simpler because they omit complex labeling steps, reducing the complexity and cost of experiments.
- Natural state of data. Without the interference of chemical labeling, label-free proteomic analysis can more naturally reflect the real state of proteins in biological samples.
- Wide applicability. For samples that cannot be chemically labeled or where labeling methods may introduce bias, label-free proteomics provides an effective analytical option.
Explore Our Label-Free Proteomics Analysis
Protein characterization and relative quantitative analysis
- We use liquid-mass spectrometry (LMS) technology to perform mass spectrometry analysis of proteins' enzymatically digested peptides. We perform relative quantification of proteins by comparing the signal intensity of corresponding peptides in different samples. At the same time, the proteins are analyzed qualitatively to determine their amino acid sequences and structural features.
- Application. It can be used to analyze plant samples of different varieties, different growth stages, or under different treatment conditions to understand the differences in protein expression and changing rules and to provide a basis for varietal improvement, growth regulation, etc.
Differential protein screening and identification
- By comparing the proteomic data of different samples, we screen out the differential proteins with significant changes in expression under specific conditions and conduct further identification and functional analysis. This includes subcellular localization of proteins, secondary structure prediction, and functional domain analysis.
- Application. In plant antiretroviral research, we can screen out proteins with changes in expression under adverse conditions such as drought, salinity, pests, and diseases, determine their roles in antiretroviral mechanisms, and provide target proteins for breeding antiretroviral varieties.
Protein interaction analysis
- We use protein interaction analysis techniques, such as affinity purification combined with mass spectrometry, to study the interaction relationship between proteins, construct protein interaction networks, and reveal the functional modules and signaling pathways of proteins in the organism.
- Application. In the study of microbial-plant interaction, the protein interactions between microbes and plant root secretions are analyzed, the mechanism of microbial promotion of plant growth is understood, and theoretical support for the development of biofertilizers and biopesticides is provided
Proteomic analysis of special samples
- We can also optimize the experimental process of protein extraction, enzymatic digestion, and mass spectrometry analysis for some special samples, such as micro samples, degraded samples, complex matrix samples, etc., to ensure that accurate and reliable proteomic data can be obtained.
- Application. In rare plant conservation research, proteomic analysis is performed on trace plant tissue samples to study the molecular mechanism of their growth, development, and adaptation to the environment and to provide technical support for conservation and breeding.
Our Service Process of Label-Free Proteomics Analysis
- Sample receipt and pre-processing. Standardized extraction of proteins and enzymatic digestion into peptides.
- Chromatographic separation and mass spectrometry. Peptides were separated by HPLC and entered mass spectrometry for MS1 (quantification) and MS/MS (identification) scanning.
- Data acquisition and quantification. Protein relative abundance was calculated.
- Bioinformatics analysis. Differential protein screening, functional enrichment, interaction network construction, mechanism modeling.
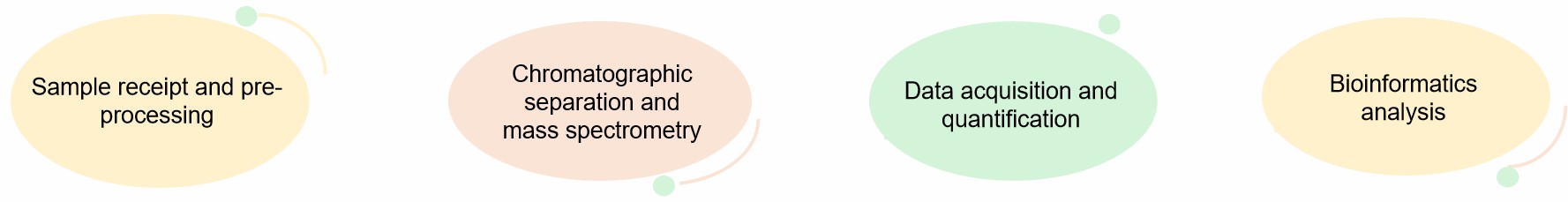 Fig.2 Process of our service. (Lifeasible)
Fig.2 Process of our service. (Lifeasible)
Highlights of Our Label-Free Proteomics Analysis
- We provide full-process services from experimental design to data interpretation, and we can also help clients customize their solutions.
- We specialize in agricultural proteomics and have successfully supported cutting-edge research on crop disease resistance and developmental regulation.
- We have a professional testing platform to ensure the reliability of data.
Lifeasible provides experience in label-free proteomics analysis. Our advanced technology platform and professional technical team aim to help you promote the research and application of proteomics in the field of agricultural research. If you are interested, please feel free to contact us.
For research or industrial raw materials, not for personal medical use!

 Fig.2 Process of our service. (Lifeasible)
Fig.2 Process of our service. (Lifeasible)