Most plant diseases are caused by a combination of lower animals, microorganisms and viruses, etc. These pathogens attack the inter-root defenses provided to the plant and cause plant diseases that result in reduced crop yields and significant economic losses. Metagenomics technology bypasses the traditional microbial isolation and culture steps to directly extract the total genomic DNA of microorganisms in the environment, providing the theoretical possibility of accessing the genetic resources available to all environmental microorganisms.

Lifeasible uses metagenomics technology to study multi-species genomes and molecular biology research tools to help detect and analyze microorganisms in plant samples. It helps you discover new resources of biocontrol microorganisms and fully understand the biocontrol microorganism-environment-pathogen interactions in the plant disease system. We obtain all the microorganisms in a particular sample by extracting nucleic acids from all the microorganisms in the plant sample, constructing sequencing libraries, sequencing them on the machine, and then quality control, assembling, comparing, and annotating the obtained data. Or we design primers based on the rDNA database, and finally, with the help of bioinformatics, we obtain information about their biodiversity and gene function.
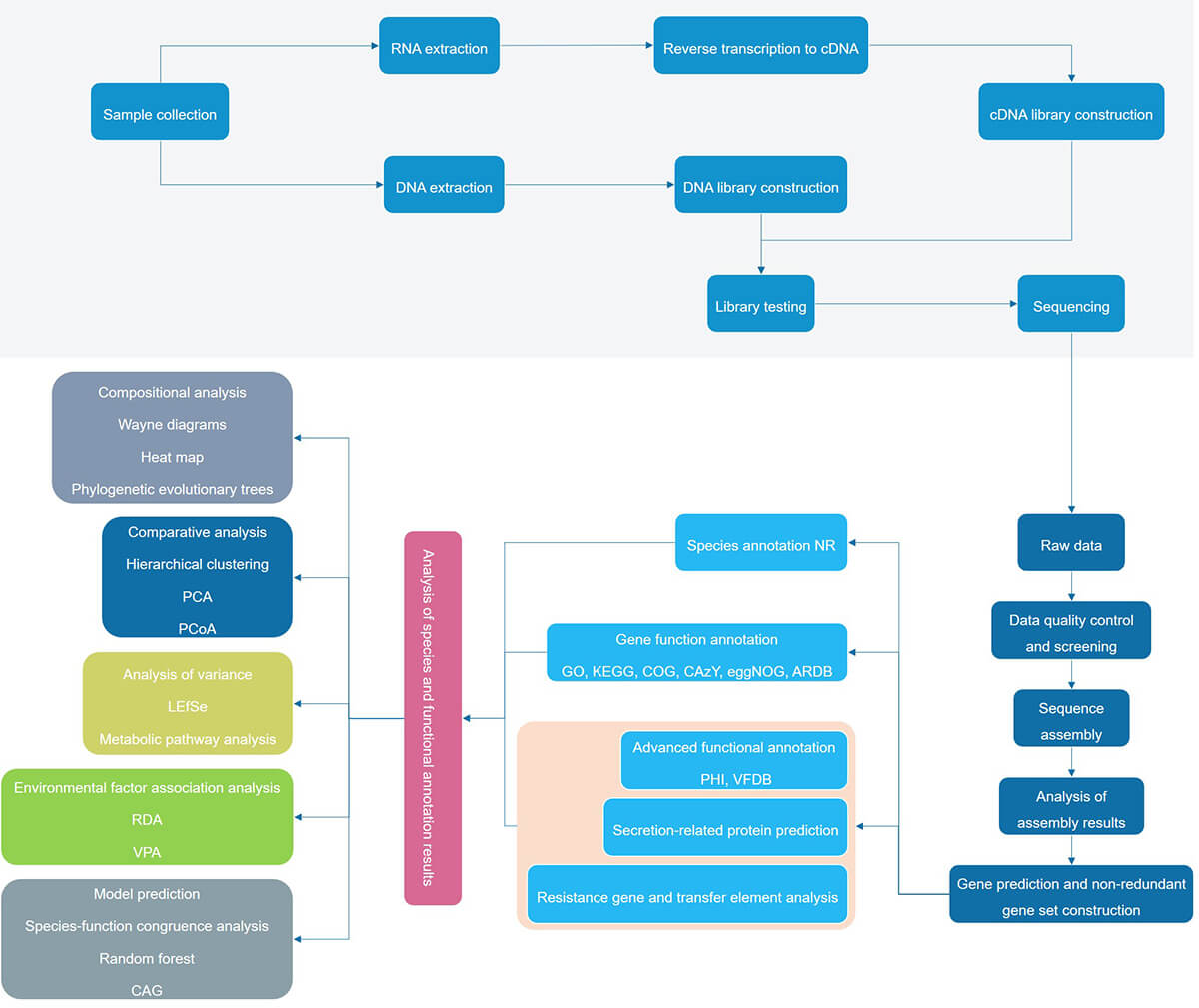 Figure 1. Metagenomics analysis process.
Figure 1. Metagenomics analysis process.
The method directly sequences all nucleic acids in the sample without bias. We use mNGS technology to detect phytopathogenic microorganisms after sample pre-treatment, nucleic acid extraction, library preparation, onboard sequencing, and meeting the quality control requirements of the test. We use specific algorithmic software to match against a dedicated in-house database of plant pathogenic microorganisms to detect possible pathogenic microbial sequences contained in the sample. This enables you to genotype organisms, identify species-level organisms, elucidate metabolic pathways, and determine the microbiota, enabling the detection of plant viruses, bacteria, fungi, parasites, and non-classical microorganisms.
|
|
Lifeasible's use of metagenomics technology opens up new avenues for the more comprehensive development of new biocontrol microbial resources and enhancing biocontrol microorganisms' inhibitory effects on pathogens. This will help you to better understand the antagonistic and promotive mechanisms between microorganisms so that you can regulate the dynamic balance between exogenous biocontrol factors and pathogens when implementing control measures and ultimately achieve plant disease control. Please feel free to contact us for more information.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Mechanisms Regulating Plant Chloroplast Biogenesis
April 15, 2025