Molecular mapping of different species of forage grasses is being performed in many research laboratories around the world using molecular markers. However, the mapping of forage grasses lags behind that of many other crops. Forage grasses are often perennial polyploids, making molecular localization a challenge. In addition, the presence of many segregating genotypes in forage grasses and the small flower size of forage grasses make it difficult to obtain segregating progeny from crosses. Little effort is currently invested in the molecular mapping of forage grasses, which hinders the development of molecular mapping forage grasses. Since most forage grasses are perennials, they can be preserved almost indefinitely as asexual reproduction plants, a distinct advantage over populations of many other crop species. Asexual propagation is particularly useful for breeders who need quantitative genes for multi-year assessment of yield or pest resistance. Therefore, it is essential to map forage grasses.
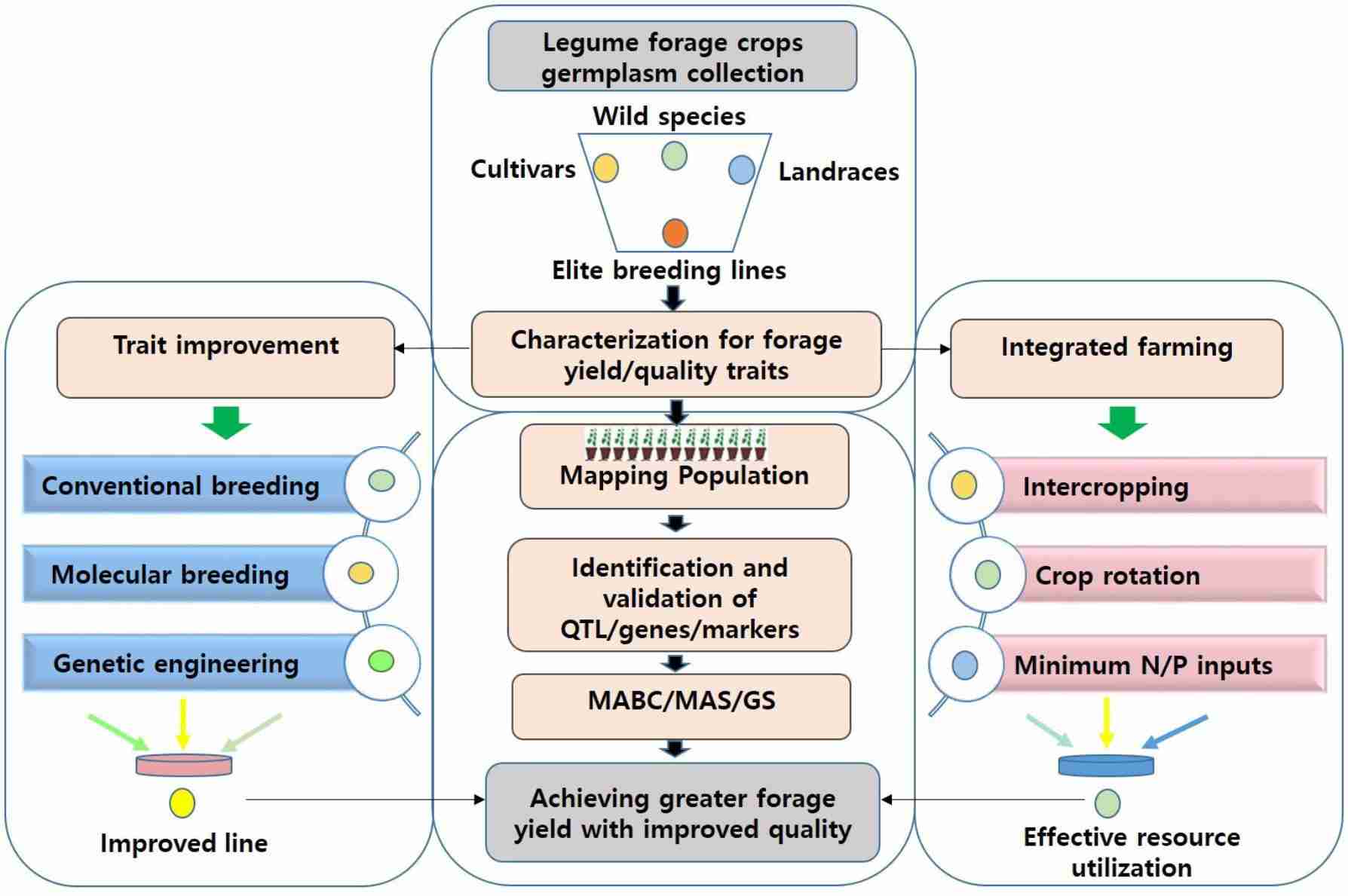 Fig.1. An integrated approach for achieving sufficient fodder and feed for livestock. (Kulkarni K P et al., 2018).
Fig.1. An integrated approach for achieving sufficient fodder and feed for livestock. (Kulkarni K P et al., 2018).
Our forage molecular biologists are developing molecular markers to map forages of several species. As an ideal partner for forage research, Lifeasible provides comprehensive molecular mapping for a wide range of forages to help clients analyze the complexity of forage genome organization and structure. We have the following cutting-edge mapping technology platforms:
We provide chromosomal DNA fragments to construct genetic maps by direct markers of chromosomal fragment segregation patterns. Restriction fragment length polymorphisms are abundant in most organisms, and an almost unlimited number can be mapped in any one crossover. Therefore, no laborious construction of marker libraries is required.
We offer random amplified polymorphic DNA markers for forage genetic mapping, a fast and simple method to collect information about a wide range of biological genetic variations. In addition, this analysis requires only a small amount of DNA to complete.
We increase the utility of PCR-based markers by developing primers for genomic regions that are more likely to show variation than randomly selected sequences. This method is the most useful genetic marker.
We offer amplified fragment length polymorphism technology to visualize sets of restriction endosomes by PCR without knowledge of the nucleotide sequence. This method allows specific co-amplification of high numbers of restriction fragments.
For accurate molecular mapping of forage grasses, we focused on parental selection and mapping populations of forage grasses:
The development of molecular mapping of forage is important because we expect to use this technology to understand better genomic relationships and identify genes for important agronomic traits. We need to do more in this area because forage grasses are critical in many parts of the world, both as livestock feed and turf. For more information or to discuss in detail, please contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Mechanisms Regulating Plant Chloroplast Biogenesis
April 15, 2025