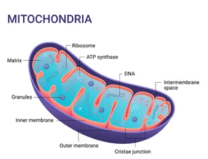
The mitochondrial genome structure of mollusks shows a high degree of variability involving many genomic features, i.e., length, gene arrangement, strand assignment, gene duplication and loss, nucleotide composition, etc. It shows extensive variation in mollusks, gastropods, and bivalves, even within the same family or genus. Mitochondrial DNA (mtDNA) has been widely used in studying biological evolution and population structure due to its unique maternal inheritance pattern and high mutation frequency. Second-generation sequencing can obtain mitochondrial DNA sequence information easily and rapidly with high throughput; therefore, molluscan mitochondrial sequencing is critical in the field of evolutionary biology research.
Mitochondria DNA sequencing is usually performed on the products of mitochondrial signature gene amplification, as well as on the smaller fragments of Gap present in the second-generation random sequencing for loop formation.
Usually, for most animal-like mitochondria and relatively small mitochondrial genomes.
a) Mainly for cases with larger mitochondrial genomes and more repetitive regions and complex structures, resulting in poor sequencing coverage, which cannot be looped using only second-generation random sequencing and cannot be Gap-completed using first-generation sequencing.
b) No high-quality mitochondrial reference genome, for the case where the reference genome of closely related species cannot be looped.
1. Description of raw sequencing data.
2. Raw sequencing data quality control.
3. Raw sequencing data quality cut and statistics.
4. Genome assembly.
5. Gene prediction.
6. rRNA/tRNA search.
7. Gene function annotation.
8. Genome circle mapping.
1. Genome-wide covariance analysis.
2. SNP, Indel detection, and annotation.
3. SV detection.
4. Phylogenetic analysis of samples.
5. SSR marker development based on sequence sequencing results.
Sequencing of Total DNA samples.
Type: non-degraded, non-contaminated DNA.
Total amount: >1 μg.
Concentration: >20 ng/μL.
The effective rate is greatly affected by sample genome size and organelle copy number, high randomness.
Suitable for reference sequences with mitochondria of closely related species.
Customers can adopt suitable sequencing protocols according to different needs; strict experimental flow from DNA extraction and quality control to library construction and up-sequencing, thus ensuring high accuracy of data. Strong analysis team; strong bioinformatics team, providing personalized bioinformatics services in addition to standard bioinformatics services.
Have completed mitochondrial/follow-up sequencing for a variety of species.
We have rich experience in personalized analysis and can choose the most suitable analysis software according to the project’s needs, only to guarantee the most accurate results.
Strict quality control processes guarantee the accuracy of results.
If you are interested in our services, or if the service you want is not listed, please feel free to contact us, and we will get back to you as soon as possible.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025