Genes are translated and modified after transcription, so gene expression products are usually proteins. Proteomics is a discipline that studies the composition, structure, and function of all proteins in the genome or cell for qualitative and quantitative analysis based on genomics and transcriptomics. Recently, proteomic analysis has been developed as a powerful tool to study the protein content of fungi, particularly basidiomycetes. The development of proteomics has facilitated mushroom research focusing on the molecular mechanisms involved in developmental stages, including extracellular and cytoplasmic effector proteins with potential or involved in the anticancer, antidiabetic, antioxidant and antibiotic synthesis, blood pressure control, vitamin and mineral supply, and other responses to environmental changes.
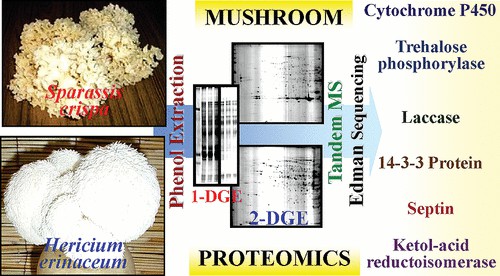 Fig. 1. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum. (Horie K, et al., 2018)
Fig. 1. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum. (Horie K, et al., 2018)
Services
Lifeasible offers comprehensive mushroom proteomics services to characterize the protein expression profile of mushrooms at different stages of growth and development and to analyze changes in protein expression between two different mushroom species. Based on proteomics data, our experts can develop strategies for studying the role of enzymes and proteins during mushroom development in future cultivation, especially in mushrooms with challenging cultivation conditions.
We have successfully performed proteomic analyses on different mushrooms such as L. rhinocerotis, T. heimii, A. bisporus, Pleurotus tuber-regium, A. cinnamomea, G. lucidum, P. ostreatus, and F. velutipes. Our mushroom proteomics services are widely used to analyze cellular metabolism, identify minerals and vitamins, and protein effectors in mushrooms, and determine quantitative changes in protein expression in response to stress exposure.
Much of Lifeasible's research on mushroom proteomics has focused on differential proteomics, which is used to screen for proteins that are differentially expressed in mushrooms in different environments, tissue morphologies, and developmental stages. We develop customized processes for mushroom differential proteomics analysis.
- Extraction and isolation of protein components from mushrooms.
- Screening and identification of differentially expressed proteins.
- Qualitative analysis of the structure, properties, and functions of differentially expressed proteins using bioinformatics.
Applications of Our Mushroom Proteomics
- Analysis of the role of enzymes and proteins in mushroom cultivation.
- Identification of developmental stages and novel mushroom compounds.
- Determining the effect of stimulants on mushroom growth and bioactive metabolite production.
- Determining quantitative changes in protein expression in response to stress.
- Investigating mushroom developmental processes.
Our ProteomicsTechnology Platforms
- Tris-saturated phenol assay and TCA/acetone precipitation.
- Two-dimensional gel electrophoresis (2-DE) or liquid chromatography coupled with mass spectrometry (LC-MS).
- Differential Gel Electrophoresis (DIGE) technique.
- High-throughput birdshot proteomics.
- Gel-free proteomics.
- iTRAQ labeling technology with 2D LC-MS/MS.
We will select the best combination of methods according to the different research objectives of our clients. Our mushroom proteomics services can be used for different purposes and have become a necessary complement to genomic and transcriptomic technologies. Our goal is to explore the intrinsic changes in mushrooms at the proteome level in different environments, tissues, and periods of growth and development. If you are interested in our services, please contact us.
Reference
- Horie K, et al. (2008) Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum provides insight into their numerous functional protein components and diversity[J]. The Journal of Proteome Research. 7(5): 1819-1835.
For research or industrial raw materials, not for personal medical use!


 Fig. 1. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum. (Horie K, et al., 2018)
Fig. 1. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum. (Horie K, et al., 2018) 
