The plant microbiome (Microbiota) refers to all microorganisms that inhabit plant hosts, including symbiotic, mutualistic symbionts, and harmful pathogens. The healthy plant microbiome includes a diverse group of microorganisms, primarily bacteria. Most root-inhabiting bacteria come from the soil. The bacterial microbial community structure of the soil, inter-, and root differed significantly and had the lowest microbial diversity within the roots.
Lifeasible continues to delve into the plant microbiome for agricultural crop production research. Not only can we grow most of the bacteria in the plant microbiome under laboratory conditions, but inoculating them in sterile plants can reproduce a natural-like community structure and deepen our understanding of plant microbes. We also provide effective solutions and technical support to enhance plant microbiome activities and to safely and effectively deploy engineered microbiota in large fields to help improve and sustain plant production.
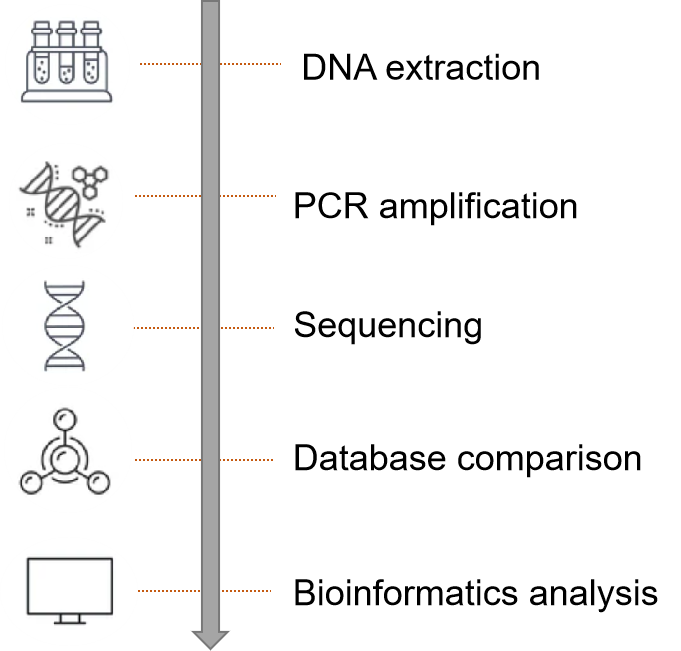
Plant microbes play an important role in influencing host growth and development, nutrient uptake, and disease resistance. Uncovering plant microbial community structure, interactions networks, and ecological functions, and how plants regulate their microbial communities in response to external stresses, can provide effective solutions to promote agricultural development.
Lifeasible is deeply involved in the field of plant science research, constantly developing new technologies and optimizing our service process to help in-depth plant research, improve agricultural productivity and promote green, healthy, and sustainable agriculture. If you are interested in us, please feel free to contact us.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025