We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Lifeasible focuses on plant-pathogen interactions and offers services to help study the role of plant proteases in plant-pathogen interactions.
The precise regulation of immune signaling is essential. Protease-mediated proteolysis represents a key form of precise regulation. Plant proteases are essential in plant immunity and regulate various immune signaling pathways, including pathogen recognition, apoplastic immunity, pattern-triggered immunity (PTI), and effector-triggered immunity (ETI). For example, some plant proteases regulate plant PTI by cleaving pattern recognition receptors (PRRs), including CERK1 and Xa21. Some proteases act directly on pathogen growth-associated proteins to inhibit pathogen growth. For example, the protein SAP1/2 secreted by Arabidopsis inhibits bacterial growth by cleaving the bacterial growth-associated protein MucD. Overall, plant proteases play a crucial role in plant-pathogen interactions (Fig. 1). In-depth understanding of the mechanisms of action of plant protease-mediated plant immunity facilitates the engineering of resistant plants.
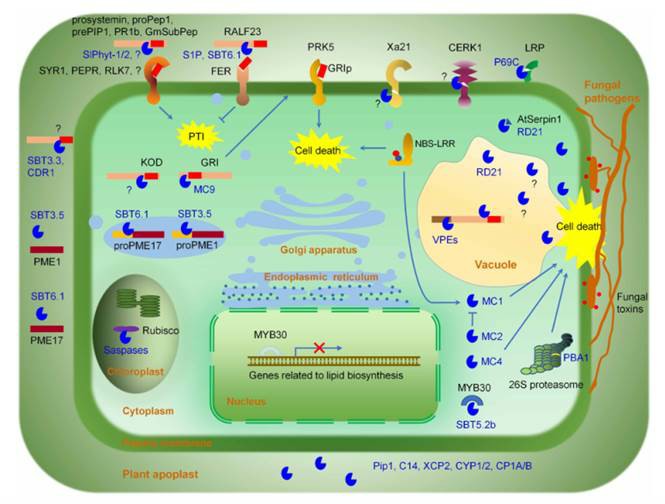 Fig. 1 Schematic of plant protease-regulated immune signaling pathways (Hou et al., 2018).
Fig. 1 Schematic of plant protease-regulated immune signaling pathways (Hou et al., 2018).
Lifeasible offers services to help explore the role of plant proteases in plant-pathogen interactions. We help explore the initial role of specific proteases in plant protection and offer further services to investigate the substrates and biochemical mechanisms of the protease.
Explore the initial role of a plant protease
We offer a preliminary exploration of the role of plant proteases in plant-pathogen interactions through knockout, knockdown, or overexpression approaches. We help to initially determine the effects of proteases on plant physical and chemical bulwarks, two types of innate immune signaling pathways (PTI and ETI), systemic acquired resistance (SAR), and the effects on invasion or growth inhibition of pathogens. We determined the effects by detecting changes in critical molecules of these affected factors.
Investigate action mechanisms and targets of a plant protease
Protein subcellular localization studies are necessary to study the function of proteins. We help determine the subcellular localization of proteases. We combine the results of initial role studies with localization information of protease to determine the general direction of the biochemical mechanism study. Plant proteases are classified into four categories (cysteine proteases, serine proteases, aspartic proteases, and metalloproteases). We then help to identify the specific class of this protease. Finally, we help investigate the targets of proteases by protein-protein interaction methods such as yeast two-hybrid (Y2H) and fluorescence resonance energy transfer (FRET). We also help explore the cleavage targets of proteases by mass spectrometry (MS).
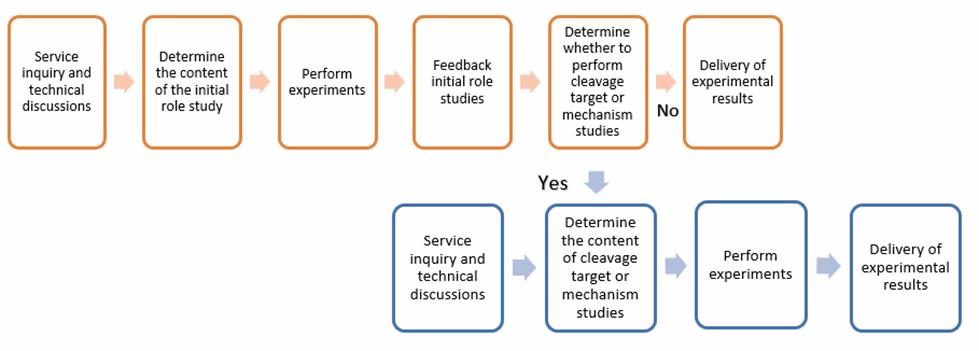
Lifeasible offers specialist plant protease research services. We help explore plant protease's initial role, specific mechanism, and cleavage targets in plant-pathogen interactions. Please contact us to customize your service.
References
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy