Telomeres are basic structures of eukaryote genomes. They distinguish natural chromosome ends from double-stranded breaks in DNA and protect chromosome ends from degradation or end-to-end fusion with other chromosomes. The telomere repeats motif (TTTAGGG)n, discovered in Arabidopsis thaliana, is the consensus telomere motif for most land plants. Although this sequence is the most common, there is also significant variation in some species. Telomere DNA sequencing is a critical step in unraveling the complexities of telomeres in plants.
Lifeasible, a leading biotechnology company specializing in plant research, has been developing innovative techniques for plant telomere DNA sequencing. With years of expertise in the industry, we have consistently provided high-quality solutions for plant scientists and researchers worldwide.
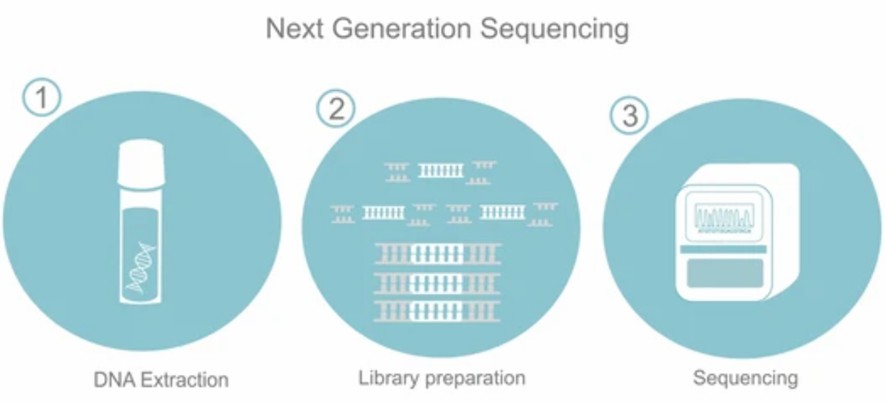 Fig. 1 Schematic diagram of the NGS process.
Fig. 1 Schematic diagram of the NGS process.
At Lifeasible, our deep understanding of plant genomics and telomere biology enables us to offer tailored solutions for diverse research needs. We leverage cutting-edge technologies and bioinformatics tools to ensure accuracy and reliability in our results. If you are interested in our services or have some questions, please feel free to contact us or make an online inquiry.
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025