Natural variation among crop varieties, local varieties, and their wild relatives provides the genetic diversity necessary for plant breeding and crop improvement. Molecular genetic studies have identified genes responsible for controlling many agriculturally essential traits. Differences in alleles selected during domestication and improvement are responsible for major differences in crop yield and other agriculturally important traits. The most valuable alleles are usually derived from local endemic varieties or related species, or even direct homologs of other plant species, and are generally caused by differences in one or more single nucleotide polymorphisms (SNPs) or insertions/deletions (indels) in specific genes. Harnessing genetic diversity and introducing superior alleles into commercial varieties has been a significant goal of crop breeding programs.
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-correlated proteins (Cas) (CRISPR/Cas) are simple, versatile, powerful, and cost-effective genome manipulation lines, which are particularly important for crop improvement. Various CRISPR/Cas toolkits have been developed to easily disrupt genes for DNA repair by non-homologous end joining (NHEJ) or double-stranded DNA breaks (DSB), and precision editing using nuclease technology has traditionally required the use of homology-directed repair (HDR). HDR is only effective at certain cell cycle stages and has different efficacies across species and cell types. HDR is only effective at certain cell cycle stages and varies in efficacy between species and cell types. Using genome editing in plant systems is particularly difficult, hampering the ability to generate precision-edited crops with desired phenotypes.
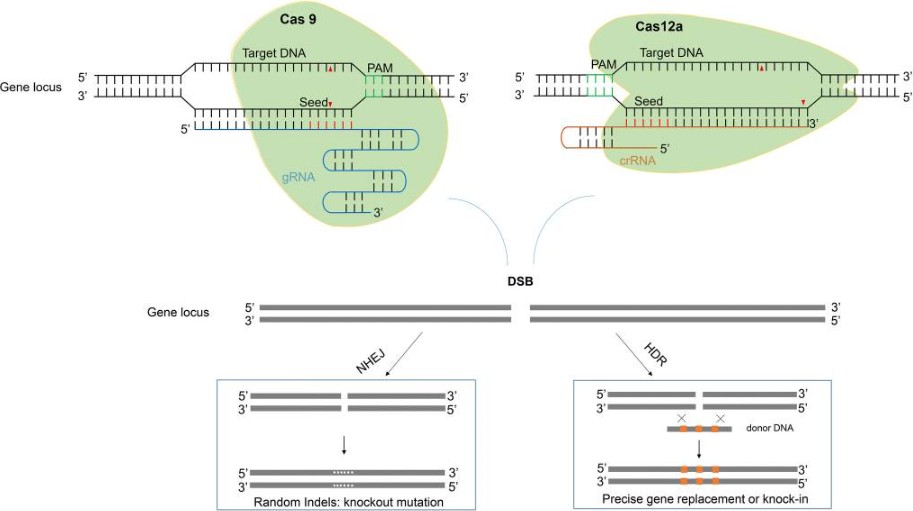 Fig. 1. Two major pathways underlying the repair of double-stranded DNA breaks induced by CRISPR/Cas. (Li et al., 2020)
Fig. 1. Two major pathways underlying the repair of double-stranded DNA breaks induced by CRISPR/Cas. (Li et al., 2020)
Lifeasible has been at the forefront of advances in plant biotechnology for many years, working to enable precision gene replacement in plants through CRISPR/Cas genome editing. We use CRISPR for allelic replacement of RNA-encoding DNA, enabling all small, precise edits (substitutions, insertions, and deletions), and are compatible with Type II and Type V CRISPR systems. We have fully optimized this technology:
Homology-directed repair (HDR) is a powerful tool for targeting alleles/gene substitutions or precise genomic modifications. However, performing HDR has been difficult, especially in plants. Our solution provides CRISPR/Cas-mediated HDR, which can introduce elite alleles into commercial lines within 2-3 generations without introducing unwanted genes or traits. We have successfully used CRISPR/Cas-mediated HDR for targeted elite allele replacement or knock-in of specific genes or markers/tags to localize the target locus.
We offer the following strategies to improve the efficiency of HDR for precise sequence replacement.
With all types of CRISPR/Cas-mediated single and multiple base substitutions, as well as small and precise insertion deletions, we can rapidly, efficiently, and economically improve crops in a user-defined manner without compromising other good agronomic traits or being weighed down by a chain of deleterious genes. Lifeasible aims to perform precise gene/allele substitutions through the CRISPR/Cas system combined with traditional breeding techniques. Allele substitution, combined with traditional breeding methods, and its application to the selection of superior varieties of multiple crops for sustainable agriculture and a better environment to ensure global food security. Contact us today to learn more about our solutions.
Reference