Lifeasible assists in applying RNA interference in plant protection and provides services to synthesize and modify small RNAs (sRNAs) for plant protection research.
RNAi interference currently used for plant protection can be divided into two categories. They are host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS). HIGS-based plant protection is mainly through plant gene manipulation (GM). For example, gene-edited plant expression of insect-specific dsRNA as a plant-incorporated protectant (PIP). SIGS-based sRNAs can be used to spray foliar, soil, or seed as a plant protectant. Fig. 1 shows the advantages and disadvantages of RNA interference based on in planta and ex planta applications. Our company customizes the synthesis and modification of sRNAs based on the actual needs of RNA interference applications and the advantages and disadvantages of in planta and ex planta applications of RNA interference.
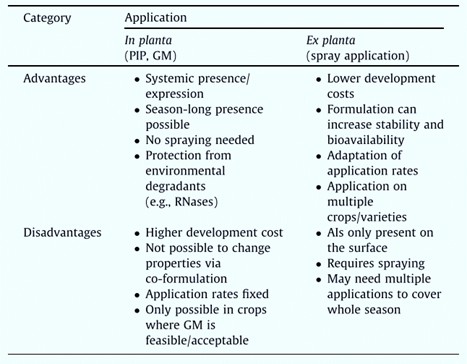 Fig. 1 Advantages and disadvantages of in planta versus ex planta application of RNA-based solutions (Bramlett et al., 2020).
Fig. 1 Advantages and disadvantages of in planta versus ex planta application of RNA-based solutions (Bramlett et al., 2020).
Synthesis of HIGS-based sRNAs
We offer services to edit plant genes to produce shRNAs or dsRNAs for plant protection. We use nanomaterials-, Agrobacterium-, electroporation-, and polyethylene glycol (PEG)-mediated genetic transformation.
Synthesis of SIGS-based sRNAs
We offer various methods for the synthesis of SIGS-based sRNAs. We provide in vitro synthesis of sRNAs, including chemical and enzymatic synthesis. We also offer in vivo biological synthesis of sRNAs, including the synthesis of sRNAs using plants, bacteria, and yeast.
Modification of sRNAs
We use chemical and biological approaches to enhance the stability of SIGS-based sRNAs to provide longer-term protection. We offer modification of SIGS-based sRNAs to enhance their uptake rate and add fluorescent tags to sRNAs to observe the uptake rate.
Efficient chemical synthesis of sRNAs
We have advanced high-throughput synthesis instruments to achieve efficient sRNA synthesis, and we use HPLC to purify chemically synthesized sRNAs. We can synthesize siRNA in one-time quantities up to gram(g) level, and our synthesis quantity can meet the needs of large-volume sRNA experiments, such as those using RNA interference to kill insects.
Economic enzymatic synthesis of sRNAs
To facilitate the enzymatic synthesis, we developed the sRNA enzymatic synthesis kit, which can meet the need of synthesizing multiple sRNAs at one time, and provide convenience for studying the plant protective effects of multiple sRNAs at one time.
Long-term biosynthesis of sRNAs
We provide specialized gene editing services for organisms such as plants, bacteria, and yeast to produce sRNAs. We help construct vectors, perform transfection, transformation or transduction, and validation of sRNAs. Gene editing-based biosynthesis can meet the needs of long-term production of sRNAs.
Chemical and biological modification of sRNAs
We offer a variety of chemical and biological methods for end modification and base modification of sRNAs. Natural RNA post-transcriptional modifications are diverse, with over 150 identified so far. Our modifications of sRNAs learn from the modifications of natural RNAs. We improve the stability of sRNA and enhance the affinity of sRNA with the enzyme Dicer by covalent alteration or isomerization of nucleotides.
Lifeasible provides sRNA synthesis and modification services. We master proven methods for gene editing in plants, bacteria, and yeast, which can be used to construct transgenic varieties to produce sRNAs. We master various methods for RNA modification, which can be used to improve the stability of sRNAs and the uptake rate of sRNAs. Please contact us for your custom synthesis of sRNAs used for plant protection.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
Get Latest Lifeasible News and Updates Directly to Your Inbox
Adaptive Evolutionary Mechanism of Plants
February 28, 2025
Unraveling Cotton Development: Insights from Multi-Omics Studies
February 27, 2025